4UIL
 
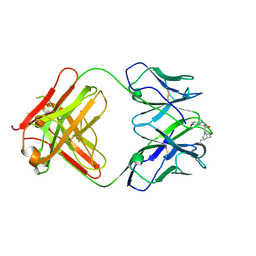 | | crystal structure of quinine-dependent Fab 314.1 with quinine | | Descriptor: | FAB 314.1, Quinine | | Authors: | Zhu, J, Zhu, J, Bougie, D.W, Aster, R.H, Springer, T.A. | | Deposit date: | 2015-03-30 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structural Basis for Quinine-Dependent Antibody Binding to Platelet Integrin Alphaiib Beta3
Blood, 126, 2015
|
|
4UIM
 
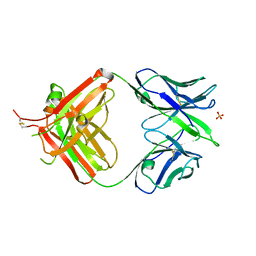 | | crystal structure of quinine-dependent Fab 314.3 | | Descriptor: | FAB 314.3, SULFATE ION | | Authors: | Zhu, J, Zhu, J, Bougie, D.W, Aster, R.H, Springer, T.A. | | Deposit date: | 2015-03-30 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Quinine-Dependent Antibody Binding to Platelet Integrin Alphaiib Beta3
Blood, 126, 2015
|
|
4UIK
 
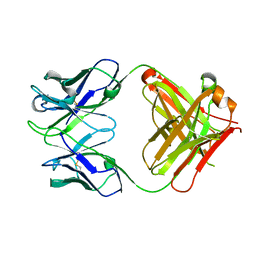 | | crystal structure of quinine-dependent Fab 314.1 | | Descriptor: | FAB 314.1 | | Authors: | Zhu, J, Zhu, J, Bougie, D.W, Aster, R.H, Springer, T.A. | | Deposit date: | 2015-03-30 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Quinine-Dependent Antibody Binding to Platelet Integrin Alphaiib Beta3
Blood, 126, 2015
|
|
4UIN
 
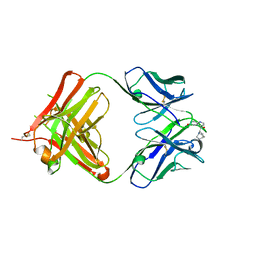 | | crystal structure of quinine-dependent Fab 314.3 with quinine | | Descriptor: | FAB 314.3, Quinine | | Authors: | Zhu, J, Zhu, J, Bougie, D.W, Aster, R.H, Springer, T.A. | | Deposit date: | 2015-03-30 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Quinine-Dependent Antibody Binding to Platelet Integrin Alphaiib Beta3
Blood, 126, 2015
|
|
4V83
 
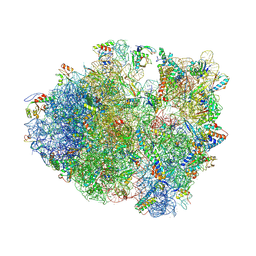 | | Crystal structure of a complex containing domain 3 from the PSIV IGR IRES RNA bound to the 70S ribosome. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhu, J, Korostelev, A, Costantino, D, Noller, H.F, Kieft, J.S. | | Deposit date: | 2010-12-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2IEL
 
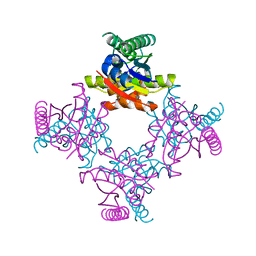 | | CRYSTAL STRUCTURE OF TT0030 from Thermus Thermophilus | | Descriptor: | Hypothetical Protein TT0030 | | Authors: | Zhu, J, Huang, J, Stepanyuk, G, Chen, L, Chang, J, Zhao, M, Xu, H, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-09-19 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF TT0030 from Thermus Thermophilus AT 1.6 ANGSTROMS RESOLUTION
To be Published
|
|
4V84
 
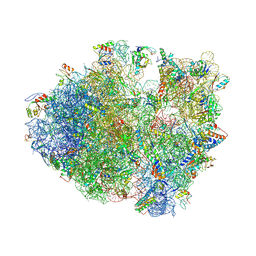 | | Crystal structure of a complex containing domain 3 of CrPV IGR IRES RNA bound to the 70S ribosome. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhu, J, Korostelev, A, Costantino, D, Noller, H.F, Kieft, J.S. | | Deposit date: | 2010-12-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6TS3
 
 | |
5MME
 
 | | Crystal structure of CREBBP bromodomain complexd with US46C | | Descriptor: | CREB-binding protein, dimethyl 5-[(5-ethanoyl-2-ethoxy-phenyl)amino]benzene-1,3-dicarboxylate | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
3FCU
 
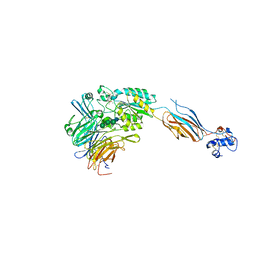 | | Structure of headpiece of integrin aIIBb3 in open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Zhu, J, Luo, B.-H, Xiao, T, Zhang, C, Nishida, N, Springer, T.A. | | Deposit date: | 2008-11-22 | | Release date: | 2009-01-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces.
Mol.Cell, 32, 2008
|
|
3FCS
 
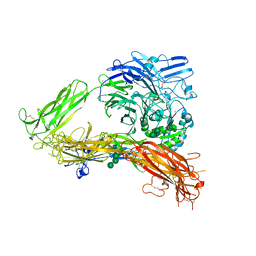 | | Structure of complete ectodomain of integrin aIIBb3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhu, J, Luo, B.-H, Xiao, T, Zhang, C, Nishida, N, Springer, T.A. | | Deposit date: | 2008-11-22 | | Release date: | 2009-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces.
Mol.Cell, 32, 2008
|
|
4YLM
 
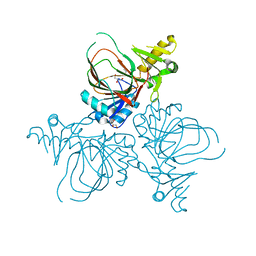 | | Structure of PvcB, an Fe, alpha-ketoglutarate dependent oxygenase from an isonitrile synthetic pathway | | Descriptor: | GLYCEROL, Pyoverdine biosynthesis protein PvcB | | Authors: | Zhu, J, Lippa, G.M, Gulick, A.M, Tipton, P.A. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Examining Reaction Specificity in PvcB, a Source of Diversity in Isonitrile-Containing Natural Products.
Biochemistry, 54, 2015
|
|
6FQO
 
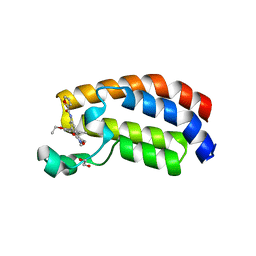 | | Crystal structure of CREBBP bromodomain complexd with DT29 | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, ~{N}-[3-(2,5-dimethyl-3-oxidanylidene-1,2-oxazol-4-yl)-5-(5-ethanoyl-2-ethoxy-phenyl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
6FQU
 
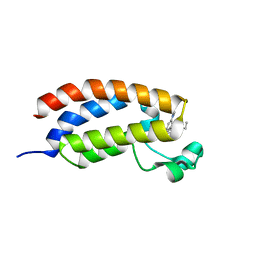 | | Crystal structure of CREBBP bromodomain complexd with DR09 | | Descriptor: | 1-[3-[3-[3,3-bis(fluoranyl)piperidin-1-yl]phenyl]-4-ethoxy-phenyl]ethanone, CREB-binding protein | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
6FR0
 
 | | Crystal structure of CREBBP bromodomain complexd with PB08 | | Descriptor: | CREB-binding protein, ~{N}-[3-(5-ethanoyl-2-ethoxy-phenyl)-5-(2-ethyl-5-methyl-3-oxidanylidene-1,2-oxazol-4-yl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-15 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
6FRF
 
 | | Crystal structure of CREBBP bromodomain complexd with PA10 | | Descriptor: | CREB-binding protein, ~{N}-[3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-(5-ethanoyl-2-ethoxy-phenyl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-15 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
7B55
 
 | | Crystal structure of CaMKII-actinin complex bound to MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-actinin-2, Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | Zhu, J, Gold, M. | | Deposit date: | 2020-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of CaMKII-actinin complex bound to MES
To Be Published
|
|
7B56
 
 | | Crystal structure of CaMKII-actinin complex bound to AMPPNP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-actinin-2, Calcium/calmodulin-dependent protein kinase type II subunit alpha, ... | | Authors: | Zhu, J, Gold, M. | | Deposit date: | 2020-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of CaMKII-actinin complex bound to MES
To Be Published
|
|
7B57
 
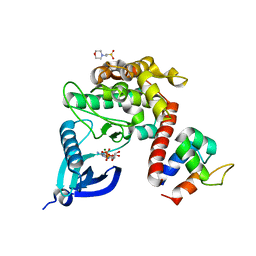 | | Crystal structure of CaMKII-actinin complex bound to ADP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Alpha-actinin-2, ... | | Authors: | Zhu, J, Gold, M. | | Deposit date: | 2020-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of CaMKII-actinin complex bound to ADP
To Be Published
|
|
6FQT
 
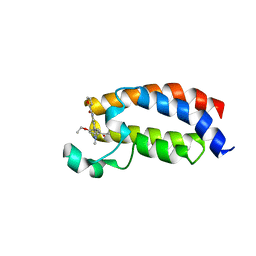 | | Crystal structure of CREBBP bromodomain complexd with DR46 | | Descriptor: | CREB-binding protein, ~{N}-[3-(5-ethanoyl-2-ethoxy-phenyl)-5-(1-methylpyrazol-3-yl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of CREBBP bromodomain complexd with DR46
To Be Published
|
|
1ZNN
 
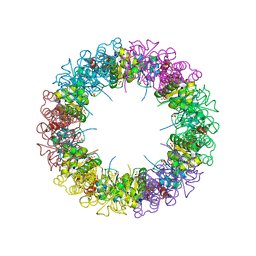 | | Structure of the synthase subunit of PLP synthase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PLP SYNTHASE, SULFATE ION | | Authors: | Zhu, J, Burgner, J.W, Harms, E, Belitsky, B.R, Smith, J.L. | | Deposit date: | 2005-05-11 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A New Arrangement of (beta/alpha)8 Barrels in the Synthase Subunit of PLP Synthase.
J.Biol.Chem., 280, 2005
|
|
5MPK
 
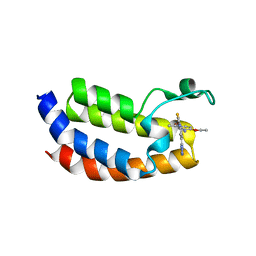 | | Crystal structure of CREBBP bromodomain complexed with DK19 | | Descriptor: | CREB-binding protein, ~{N}-(5-ethanoyl-2-ethoxy-phenyl)-3-(2~{H}-1,2,3,4-tetrazol-5-yl)-5-(1,3-thiazol-4-yl)benzamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
5MPN
 
 | | Crystal structure of CREBBP bromodomain complexed with FA26 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-ethoxy-3-[3-(2~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]ethanone, CREB-binding protein | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
5MMG
 
 | | Crystal structure of CREBBP bromodomain complexed with UT07C | | Descriptor: | 1-[4-ethoxy-3-[(1-methylsulfonylindol-6-yl)amino]phenyl]ethanone, CREB-binding protein | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
5OWK
 
 | |
