139D
 
 | |
134D
 
 | |
1HWV
 
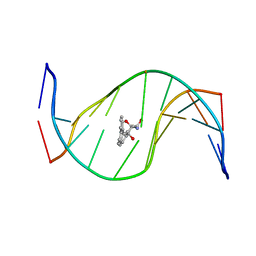
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1S)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-10 | | Release date: | 2001-03-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
1HX4
 
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1R)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-03-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
4IUR
 
 | |
6WXY
 
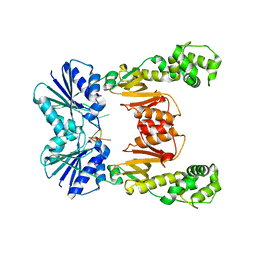 | | crystal structure of cA6-bound Card1 | | Descriptor: | Card1, cA6 | | Authors: | Rostol, J, Xie, W, Patel, D.J, Marraffini, L. | | Deposit date: | 2020-05-12 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Card1 nuclease provides defence during type III CRISPR immunity.
Nature, 590, 2021
|
|
3KHR
 
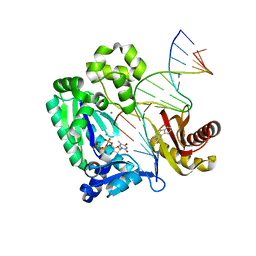 | | Dpo4 post-extension ternary complex with the correct C opposite the 2-aminofluorene-guanine [AF]G lesion | | Descriptor: | 2-AMINOFLUORENE, 5'-D(*CP*C*TP*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*CP*(DDG))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-10-30 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mechanism of error-free and semitargeted mutagenic bypass of an aromatic amine lesion by Y-family polymerase Dpo4.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV0
 
 | | Crystal structure of HET-C2: A FUNGAL GLYCOLIPID TRANSFER PROTEIN (GLTP) | | Descriptor: | HET-C2 | | Authors: | Simanshu, D.K, Kenoth, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2009-11-28 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determination and tryptophan fluorescence of heterokaryon incompatibility C2 protein (HET-C2), a fungal glycolipid transfer protein (GLTP), provide novel insights into glycolipid specificity and membrane interaction by the GLTP fold.
J.Biol.Chem., 285, 2010
|
|
5DEA
 
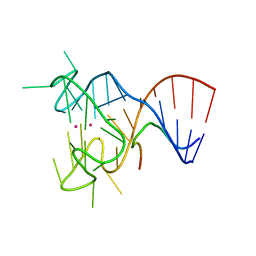 | | Crystal structure of the complex between human FMRP RGG motif and G-quadruplex RNA, cesium bound form. | | Descriptor: | CESIUM ION, Fragile X mental retardation protein 1, POTASSIUM ION, ... | | Authors: | Vasilyev, N, Polonskaia, A, Darnell, J.C, Darnell, R.B, Patel, D.J, Serganov, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7973 Å) | | Cite: | Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5DET
 
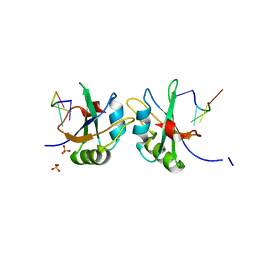 | | X-ray structure of human RBPMS in complex with the RNA | | Descriptor: | RNA (5'-R(*UP*CP*AP*C)-3'), RNA (5'-R(P*UP*CP*AP*CP*U)-3'), RNA-binding protein with multiple splicing, ... | | Authors: | Teplova, M, Farazi, T.A, Tuschl, T, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis underlying CAC RNA recognition by the RRM domain of dimeric RNA-binding protein RBPMS.
Q. Rev. Biophys., 49, 2016
|
|
5DAH
 
 | |
5DAG
 
 | |
5DDQ
 
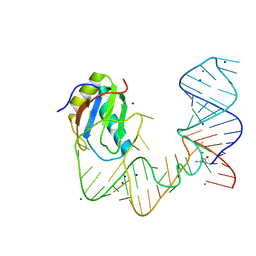 | | L-glutamine riboswitch bound with L-glutamine soaked with Mn2+ | | Descriptor: | GLUTAMINE, L-glutamine riboswitch RNA (61-MER), MAGNESIUM ION, ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
3U5O
 
 | |
3U5M
 
 | |
4ENB
 
 | | Crystal structure of fluoride riboswitch, bound to Iridium | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, IRIDIUM HEXAMMINE ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
4ENC
 
 | | Crystal structure of fluoride riboswitch | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, MAGNESIUM ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
4EZH
 
 | |
2EVS
 
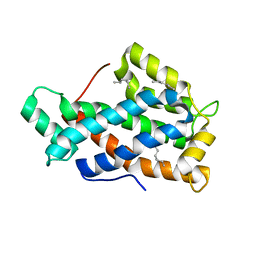 | | Crystal structure of human Glycolipid Transfer Protein complexed with n-hexyl-beta-D-glucoside | | Descriptor: | DECANE, Glycolipid transfer protein, HEXANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2EUK
 
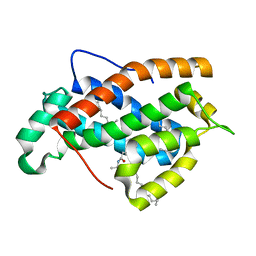 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with 24:1 Galactosylceramide | | Descriptor: | (15E)-TETRACOS-15-ENOIC ACID, Glycolipid transfer protein, N-OCTANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2EVT
 
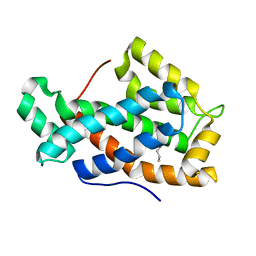 | | Crystal structure of D48V mutant of human Glycolipid Transfer Protein | | Descriptor: | Glycolipid transfer protein, HEXANE | | Authors: | Malinina, L, Malakhova, M.L, Teplov, A, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2F8T
 
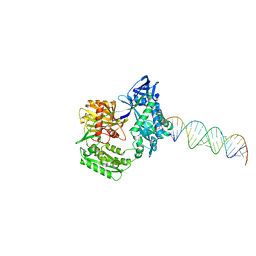 | |
2F6J
 
 | |
2FSA
 
 | |
4NTO
 
 | | Crystal structure of D60A mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C2 ceramide-1-phosphate (d18:1/2:0) at 2.15 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, DI(HYDROXYETHYL)ETHER, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
