3KCU
 
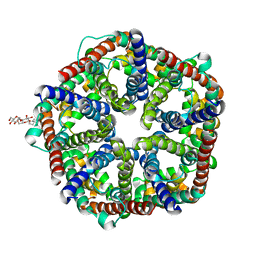 | | Structure of formate channel | | Descriptor: | 2-(6-(2-CYCLOHEXYLETHOXY)-TETRAHYDRO-4,5-DIHYDROXY-2(HYDROXYMETHYL)-2H-PYRAN-3-YLOXY)-TETRAHYDRO-6(HYDROXYMETHYL)-2H-PY RAN-3,4,5-TRIOL, Probable formate transporter 1 | | Authors: | Wang, Y, Huang, Y, Wang, J, Yan, N, Shi, Y. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel
Nature, 462, 2009
|
|
3KCV
 
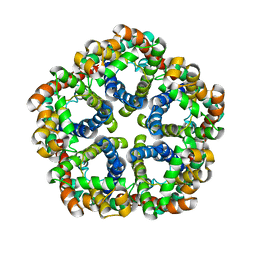 | | Structure of formate channel | | Descriptor: | Probable formate transporter 1 | | Authors: | Wang, Y, Huang, Y, Wang, J, Yan, N, Shi, Y. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel
Nature, 462, 2009
|
|
5KQE
 
 | |
6ON1
 
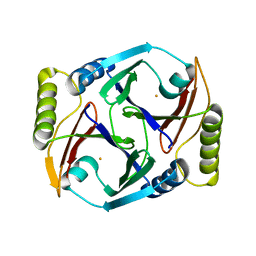 | | A resting state structure of L-DOPA dioxygenase from Streptomyces sclerotialus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, L-DOPA dioxygenase | | Authors: | Wang, Y, Shin, I, Fu, Y, Colabroy, K, Liu, A. | | Deposit date: | 2019-04-19 | | Release date: | 2019-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Crystal Structures of L-DOPA Dioxygenase fromStreptomyces sclerotialus.
Biochemistry, 58, 2019
|
|
1T8P
 
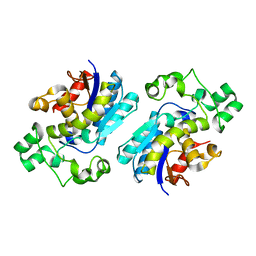 | | Crystal structure of Human erythrocyte 2,3-bisphosphoglycerate mutase | | Descriptor: | Bisphosphoglycerate mutase | | Authors: | Wang, Y, Wei, Z, Bian, Q, Cheng, Z, Wan, M, Liu, L, Gong, W. | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human bisphosphoglycerate mutase
J.Biol.Chem., 279, 2004
|
|
3K5Q
 
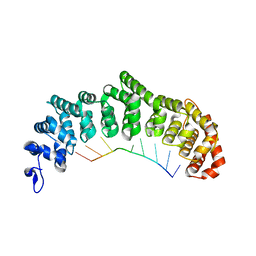 | | Crystal structure of FBF-2/FBE complex | | Descriptor: | 5'-R(P*UP*GP*UP*AP*CP*UP*AP*UP*A)-3', Fem-3 mRNA-binding factor 2 | | Authors: | Wang, Y, Opperman, L, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3K5Z
 
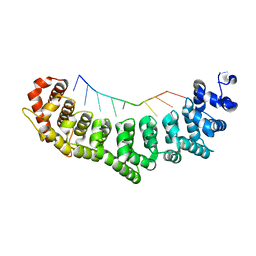 | | Crystal structure of FBF-2/gld-1 FBEa G4A mutant complex | | Descriptor: | 5'-R(*UP*GP*UP*AP*CP*CP*AP*UP*A)-3', Fem-3 mRNA-binding factor 2 | | Authors: | Wang, Y, Opperman, L, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3K64
 
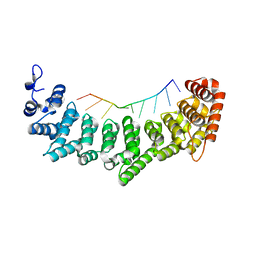 | | Crystal structure of FBF-2/fem-3 PME complex | | Descriptor: | 5'-R(*UP*GP*UP*GP*UP*CP*AP*UP*U)-3', Fem-3 mRNA-binding factor 2 | | Authors: | Wang, Y, Opperman, L, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6VI8
 
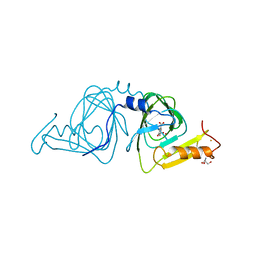 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a superoxo bound structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VIA
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a seven-membered lactone bound structure | | Descriptor: | (2R,3E)-2-hydroxy-3-imino-2,3-dihydrooxepine-4-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4M8M
 
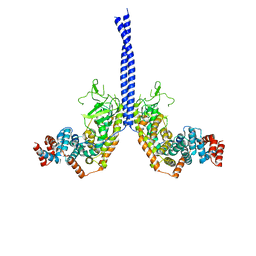 | |
4X3O
 
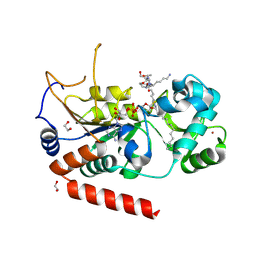 | | Sirt2 in complex with a myristoyl peptide | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | Authors: | Wang, Y, Zhang, W.Z, Hao, Q. | | Deposit date: | 2014-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deacylation Mechanism by SIRT2 Revealed in the 1'-SH-2'-O-Myristoyl Intermediate Structure.
Cell Chem Biol, 24, 2017
|
|
4X3P
 
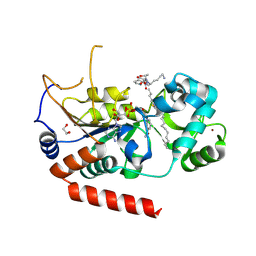 | | Sirt2 in complex with a myristoyl peptide | | Descriptor: | 1,2-ETHANEDIOL, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, Y, Zhang, W, Hao, Q. | | Deposit date: | 2014-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deacylation Mechanism by SIRT2 Revealed in the 1'-SH-2'-O-Myristoyl Intermediate Structure.
Cell Chem Biol, 24, 2017
|
|
8D5N
 
 | | Crystal structure of Ld-HF10 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2022-06-05 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide Centric V beta Specific Germline Contacts Shape a Specialist T Cell Response.
Front Immunol, 13, 2022
|
|
8D5P
 
 | | Mouse TCR TG6 | | Descriptor: | TCR-alpha, TCR-beta | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2022-06-05 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Peptide Centric V beta Specific Germline Contacts Shape a Specialist T Cell Response.
Front Immunol, 13, 2022
|
|
8D5Q
 
 | | TCR TG6 in complex with Ld-HF10 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dense granule protein 6, HF10 peptide, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2022-06-05 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Peptide Centric V beta Specific Germline Contacts Shape a Specialist T Cell Response.
Front Immunol, 13, 2022
|
|
6XJV
 
 | | MCU holocomplex in High-calcium state | | Descriptor: | Calcium uniporter protein, mitochondrial, Calcium uptake protein 1, ... | | Authors: | Wang, Y, Jiang, Y. | | Deposit date: | 2020-06-24 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structural insights into the Ca 2+ -dependent gating of the human mitochondrial calcium uniporter.
Elife, 9, 2020
|
|
5IJC
 
 | | The crystal structure of mouse TLR4/MD-2/neoseptin-3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IJB
 
 | | The ligand-free structure of the mouse TLR4/MD-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IJD
 
 | | The crystal structure of mouse TLR4/MD-2/lipid A complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3GX8
 
 | | Structural and biochemical characterization of yeast monothiol glutaredoxin Grx5 | | Descriptor: | Monothiol glutaredoxin-5, mitochondrial, SULFATE ION | | Authors: | Wang, Y, He, Y.X, Yu, J, Xiong, Y, Chen, Y, Zhou, C.Z. | | Deposit date: | 2009-04-01 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Structural and biochemical characterization of yeast monothiol glutaredoxin Grx5
To be Published
|
|
4ZHQ
 
 | | Crystal structure of Tubulin-Stathmin-TTL-MMAE Complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Zhang, R. | | Deposit date: | 2015-04-26 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules
Mol.Pharmacol., 89, 2016
|
|
4ZI7
 
 | | CRYSTAL STRUCTURE OF TUBULIN-STATHMIN-TTL-HTI286 COMPLEX | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Zhang, R. | | Deposit date: | 2015-04-27 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules
Mol.Pharmacol., 89, 2016
|
|
4ZOL
 
 | | Crystal Structure of Tubulin-Stathmin-TTL-Tubulysin M Complex | | Descriptor: | (2R,4R)-4-{[(2-{(1R,3R)-1-(acetyloxy)-4-methyl-3-[methyl(N-{[(2S)-1-methylpiperidin-2-yl]carbonyl}-D-isoleucyl)amino]pentyl}-1,3-thiazol-4-yl)carbonyl]amino}-2-methyl-5-phenylpentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Zhang, R. | | Deposit date: | 2015-05-06 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules
Mol.Pharmacol., 89, 2016
|
|
5BMV
 
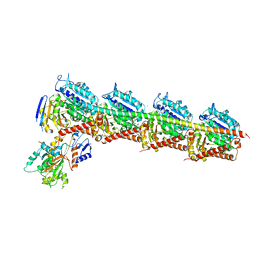 | | CRYSTAL STRUCTURE OF TUBULIN-STATHMIN-TTL-Vinblastine COMPLEX | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Chen, Q, Zhang, R. | | Deposit date: | 2015-05-23 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules.
Mol.Pharmacol., 89, 2016
|
|
