5F59
 
 | | The crystal structure of MLL3 SET domain | | Descriptor: | Histone-lysine N-methyltransferase 2C, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-04 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
5F6L
 
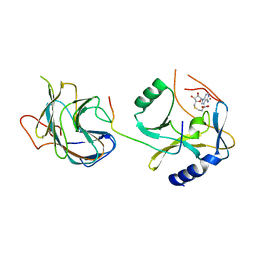 | | The crystal structure of MLL1 (N3861I/Q3867L) in complex with RbBP5 and Ash2L | | Descriptor: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
5F5E
 
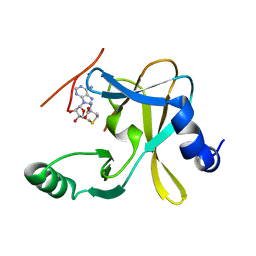 | |
5F6K
 
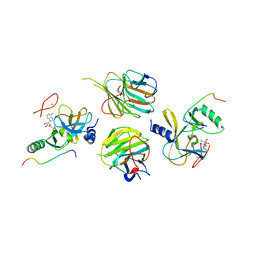 | | Crystal structure of the MLL3-Ash2L-RbBP5 complex | | Descriptor: | Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
1ZGL
 
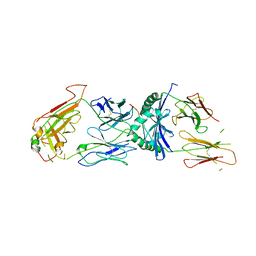 | | Crystal structure of 3A6 TCR bound to MBP/HLA-DR2a | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, Myelin basic protein, ... | | Authors: | Li, Y, Huang, Y, Lue, J, Quandt, J.A, Martin, R, Mariuzza, R.A. | | Deposit date: | 2005-04-21 | | Release date: | 2005-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule.
Embo J., 24, 2005
|
|
4G72
 
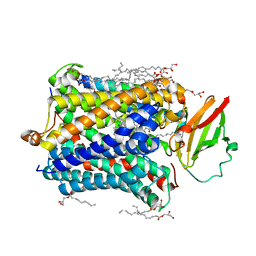 | |
4G7Q
 
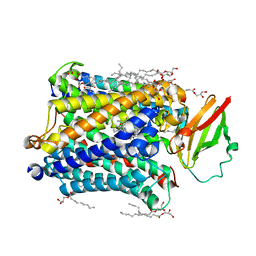 | |
4G7R
 
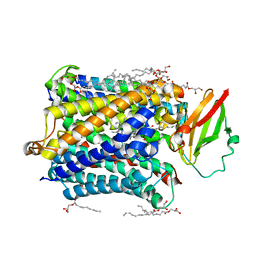 | |
5FNA
 
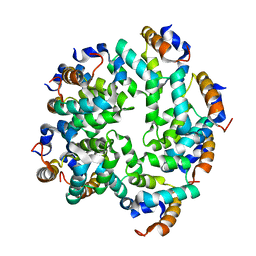 | | Cryo-EM reconstruction of caspase-1 CARD | | Descriptor: | Caspase-1 | | Authors: | Li, Y, Lu, A, Schmidt, F.I, Yin, Q, Chen, S, Fu, T.M, Tong, A.B, Ploegh, H.L, Mao, Y, Wu, H. | | Deposit date: | 2015-11-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular Basis of Caspase-1 Polymerization and its Inhibition by a Novel Capping Mechanism
Nat.Struct.Mol.Biol., 23, 2016
|
|
4N4Y
 
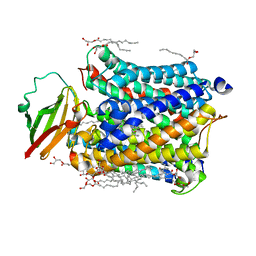 | |
6N1I
 
 | | Cryo-EM structure of NLRC4-CARD filament | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Li, Y, Fu, T, Wu, H. | | Deposit date: | 2018-11-08 | | Release date: | 2018-12-05 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3M5D
 
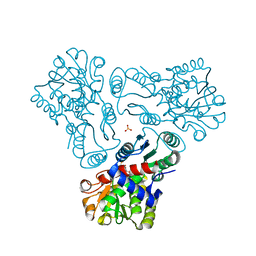 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302R mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
3M5C
 
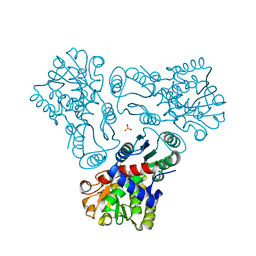 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302E mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
3N54
 
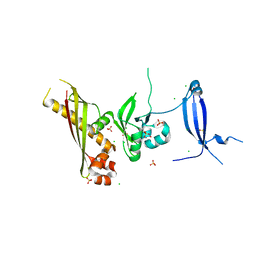 | | Crystal Structure of the GerBC protein | | Descriptor: | CHLORIDE ION, SULFATE ION, Spore germination protein B3 | | Authors: | Li, Y, Setlow, B, Setlow, P, Hao, B. | | Deposit date: | 2010-05-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the GerBC Component of a Bacillus subtilis Spore Germinant Receptor.
J.Mol.Biol., 402, 2010
|
|
8JOL
 
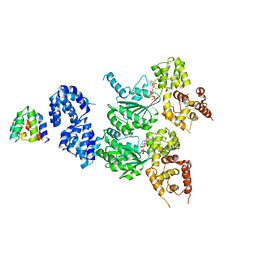 | | cryo-EM structure of the CED-4/CED-3 holoenzyme | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 3, Cell death protein 4, ... | | Authors: | Li, Y, Tian, L, Zhang, Y, Shi, Y. | | Deposit date: | 2023-06-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into CED-3 activation.
Life Sci Alliance, 6, 2023
|
|
8JO0
 
 | |
8JNS
 
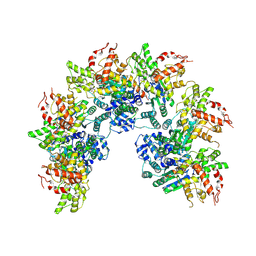 | | cryo-EM structure of a CED-4 hexamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | Authors: | Li, Y, Shi, Y. | | Deposit date: | 2023-06-06 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into CED-3 activation.
Life Sci Alliance, 6, 2023
|
|
7YTC
 
 | | Cryo-EM structure of human FcmR bound to IgM-Fc/J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, Immunoglobulin J chain, ... | | Authors: | Li, Y, Shen, H, Xiao, J. | | Deposit date: | 2022-08-14 | | Release date: | 2023-02-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Immunoglobulin M perception by Fc mu R.
Nature, 615, 2023
|
|
7YTD
 
 | | Cryo-EM structure of four human FcmR bound to IgM-Fc/J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, Immunoglobulin J chain, ... | | Authors: | Li, Y, Shen, H, Xiao, J. | | Deposit date: | 2022-08-14 | | Release date: | 2023-02-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Immunoglobulin M perception by Fc mu R.
Nature, 615, 2023
|
|
7YTE
 
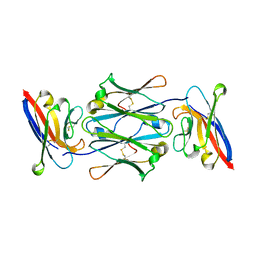 | |
7YSG
 
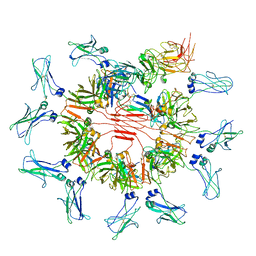 | | Cryo-EM structure of human FcmR bound to sIgM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Li, Y, Shen, H, Xiao, J. | | Deposit date: | 2022-08-12 | | Release date: | 2023-02-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Immunoglobulin M perception by Fc mu R.
Nature, 615, 2023
|
|
1HQR
 
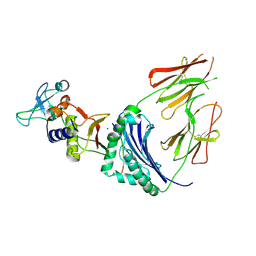 | | CRYSTAL STRUCTURE OF A SUPERANTIGEN BOUND TO THE HIGH-AFFINITY, ZINC-DEPENDENT SITE ON MHC CLASS II | | Descriptor: | HLA-DR ALPHA CHAIN, HLA-DR BETA CHAIN, MYELIN BASIC PROTEIN, ... | | Authors: | Li, Y, Li, H, Dimasi, N, Schlievert, P, Mariuzza, R. | | Deposit date: | 2000-12-19 | | Release date: | 2001-01-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a superantigen bound to the high-affinity, zinc-dependent site on MHC class II.
Immunity, 14, 2001
|
|
8W8F
 
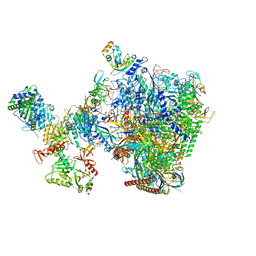 | | human co-transcriptional RNA capping enzyme RNGTT-CMTR1 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (36-MER), DNA (45-MER), ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
8W8E
 
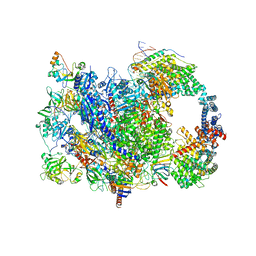 | | human co-transcriptional RNA capping enzyme RNGTT | | Descriptor: | DNA (36-MER), DNA (45-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
4XVJ
 
 | |
