1ZWW
 
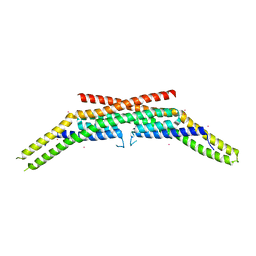 | | Crystal structure of endophilin-A1 BAR domain | | Descriptor: | CADMIUM ION, SH3-containing GRB2-like protein 2 | | Authors: | Weissenhorn, W. | | Deposit date: | 2005-06-06 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Endophilin-A1 BAR Domain.
J.Mol.Biol., 351, 2005
|
|
1ENV
 
 | | ATOMIC STRUCTURE OF THE ECTODOMAIN FROM HIV-1 GP41 | | Descriptor: | HIV-1 ENVELOPE PROTEIN CHIMERA CONSISTING OF A FRAGMENT OF GCN4 ZIPPER CLONED N-TERMINAL TO TWO FRAGMENTS OF GP41 | | Authors: | Weissenhorn, W, Dessen, A, Harrison, S.C, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1997-06-27 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Atomic structure of the ectodomain from HIV-1 gp41.
Nature, 387, 1997
|
|
1EBO
 
 | | CRYSTAL STRUCTURE OF THE EBOLA VIRUS MEMBRANE-FUSION SUBUNIT, GP2, FROM THE ENVELOPE GLYCOPROTEIN ECTODOMAIN | | Descriptor: | CHLORIDE ION, EBOLA VIRUS ENVELOPE PROTEIN CHIMERA CONSISTING OF A FRAGMENT OF GCN4 ZIPPER CLONED N-TERMINAL TO A FRAGMENT OF GP2, ZINC ION | | Authors: | Weissenhorn, W, Carfi, A, Lee, K.H, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1998-11-03 | | Release date: | 1999-07-02 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain.
Mol.Cell, 2, 1998
|
|
1XB4
 
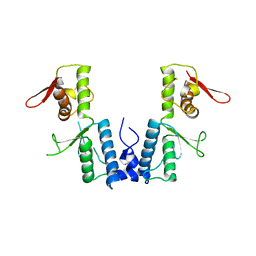 | |
7Q1Z
 
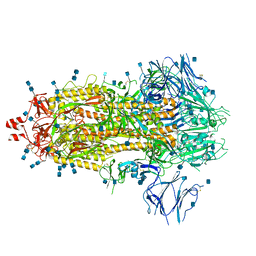 | | Structure of formaldehyde cross-linked SARS-CoV-2 S glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Sulbaran, G, Effantin, G, Schoehn, G, Weissenhorn, W. | | Deposit date: | 2021-10-22 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Immunization with synthetic SARS-CoV-2 S glycoprotein virus-like particles protects macaques from infection.
Cell Rep Med, 3, 2022
|
|
1H2C
 
 | | Ebola virus matrix protein VP40 N-terminal domain in complex with RNA (High-resolution VP40[55-194] variant). | | Descriptor: | 5'-R(*UP*GP*AP)-3', MATRIX PROTEIN VP40 | | Authors: | Gomis-Ruth, F.X, Dessen, A, Bracher, A, Klenk, H.D, Weissenhorn, W. | | Deposit date: | 2002-08-05 | | Release date: | 2003-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Matrix Protein Vp40 from Ebola Virus Octamerizes Into Pore-Like Structures with Specific RNA Binding Properties
Structure, 11, 2003
|
|
1H2D
 
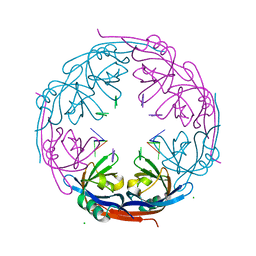 | | Ebola virus matrix protein VP40 N-terminal domain in complex with RNA (Low-resolution VP40[31-212] variant). | | Descriptor: | 5'-R(*UP*GP*AP)-3', CHLORIDE ION, MATRIX PROTEIN VP40 | | Authors: | Gomis-Ruth, F.X, Dessen, A, Bracher, A, Klenk, H.D, Weissenhorn, W. | | Deposit date: | 2002-08-06 | | Release date: | 2003-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Matrix Protein Vp40 from Ebola Virus Octamerizes Into Pore-Like Structures with Specific RNA Binding Properties
Structure, 11, 2003
|
|
2XZE
 
 | |
1MQS
 
 | |
2XA3
 
 | | crystal structure of the broadly neutralizing llama VHH D7 and its mode of HIV-1 gp120 interaction | | Descriptor: | LLAMA HEAVY CHAIN ANTIBODY D7, SULFATE ION | | Authors: | Hinz, A, Lutje Hulsik, D, Forsman, A, Koh, W, Belrhali, H, Gorlani, A, de Haard, H, Weiss, R.A, Verrips, T, Weissenhorn, W. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the neutralizing Llama V(HH) D7 and its mode of HIV-1 gp120 interaction.
PLoS ONE, 5, 2010
|
|
7AEJ
 
 | |
5O2U
 
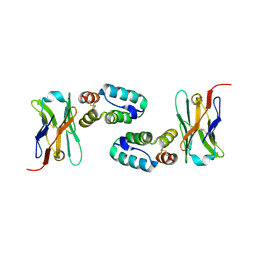 | | Llama VHH in complex with p24 | | Descriptor: | Capsid protein p24, VHH 59H10 | | Authors: | Caillat, C, Verrips, T, Weissenhorn, W. | | Deposit date: | 2017-05-22 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Unravelling the Molecular Basis of High Affinity Nanobodies against HIV p24: In Vitro Functional, Structural, and in Silico Insights.
ACS Infect Dis, 3, 2017
|
|
2XRA
 
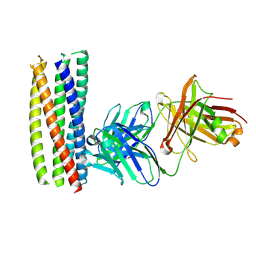 | | crystal structure of the HK20 Fab in complex with a gp41 mimetic 5- Helix | | Descriptor: | HK20, HUMAN MONOCLONAL ANTIBODY HEAVY CHAIN, HUMAN MONOCLONAL ANTIBODY LIGHT CHAIN, ... | | Authors: | Sabin, C, Corti, D, Buzon, V, Seaman, M.S, Lutje Hulsik, D, Hinz, A, Vanzetta, F, Agatic, G, Silacci, C, Langedijk, J.P.M, Mainetti, L, Scarlatti, G, Sallusto, F, Weiss, R, Lanzavecchia, A, Weissenhorn, W. | | Deposit date: | 2010-09-13 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and size-dependent neutralization properties of HK20, a human monoclonal antibody binding to the highly conserved heptad repeat 1 of gp41.
PLoS Pathog., 6, 2010
|
|
6HIG
 
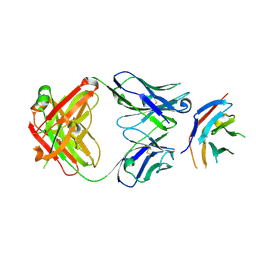 | | hPD-1/NBO1a Fab complex | | Descriptor: | Heavy Chain, Light Chain, Programmed cell death protein 1 | | Authors: | Loredo-Varela, J.L, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | Deposit date: | 2018-08-29 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tumor suppression of novel anti-PD-1 antibodies mediated through CD28 costimulatory pathway.
J.Exp.Med., 216, 2019
|
|
5M1M
 
 | |
7ZCH
 
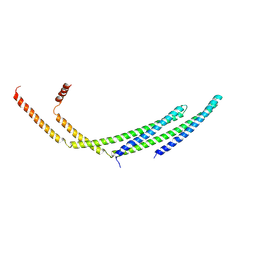 | | CHMP2A-CHMP3 heterodimer (410 Angstrom diameter) | | Descriptor: | Charged multivesicular body protein 2a, Charged multivesicular body protein 3 | | Authors: | Azad, K, Desfosses, A, Effantin, G, Schoehn, G, Weissenhorn, W. | | Deposit date: | 2022-03-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of CHMP2A-CHMP3 ESCRT-III polymer assembly and membrane cleavage.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZCG
 
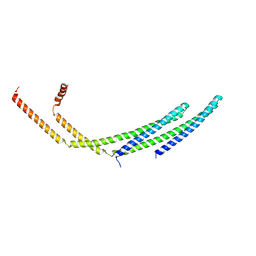 | | CHMP2A-CHMP3 heterodimer (430 Angstrom diameter) | | Descriptor: | Charged multivesicular body protein 2a, Charged multivesicular body protein 3 | | Authors: | Azad, K, Desfosses, A, Effantin, G, Schoehn, G, Weissenhorn, W. | | Deposit date: | 2022-03-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of CHMP2A-CHMP3 ESCRT-III polymer assembly and membrane cleavage.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5HM1
 
 | |
7AJ6
 
 | |
1FVH
 
 | |
1FVF
 
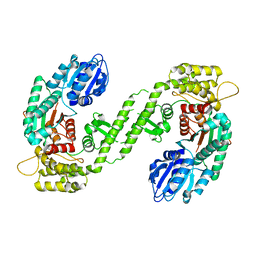 | |
2X7A
 
 | | Structural basis of HIV-1 tethering to membranes by the Bst2-tetherin ectodomain | | Descriptor: | BONE MARROW STROMAL ANTIGEN 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Natrajan, G, McCarthy, A.A, Weissenhorn, W. | | Deposit date: | 2010-02-25 | | Release date: | 2010-04-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Basis of HIV-1 Tethering to Membranes by the Bst-2/Tetherin Ectodomain.
Cell Host Microbe, 7, 2010
|
|
2X7R
 
 | |
8FFR
 
 | | Revised structure of the rabies virus nucleoprotein-RNA complex | | Descriptor: | Nucleoprotein, PHOSPHATE ION, RNA (99-MER) | | Authors: | Leyrat, C, Bourhis, J.M, Albertini, A.A.V, Wernimont, A.K, Muziol, T, Ravelli, R.B.G, Weissenhorn, W, Ruigrok, R.W.H, Jamin, M. | | Deposit date: | 2022-12-09 | | Release date: | 2023-01-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure and Dynamics of the Unassembled Nucleoprotein of Rabies Virus in Complex with Its Phosphoprotein Chaperone Module.
Viruses, 14, 2022
|
|
4D81
 
 | | Metallosphera sedula Vps4 crystal structure | | Descriptor: | AAA ATPase, central domain protein, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Caillat, C, Macheboeuf, P, Wu, Y, McCarthy, A.A, Boeri-Erba, E, Effantin, G, Gottlinger, H.G, Weissenhorn, W, Renesto, P. | | Deposit date: | 2014-12-02 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Asymmetric Ring Structure of Vps4 Required for Escrt-III Disassembly.
Nat.Commun., 6, 2015
|
|
