1SPS
 
 | |
1SHB
 
 | |
1SHA
 
 | |
1TDF
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI THIOREDOXIN REDUCTASE REFINED AT 2 ANGSTROM RESOLUTION: IMPLICATIONS FOR A LARGE CONFORMATIONAL CHANGE DURING CATALYSIS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOREDOXIN REDUCTASE | | Authors: | Waksman, G, Krishna, T.S.R, Williams Junior, C.H, Kuriyan, J. | | Deposit date: | 1994-01-14 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Escherichia coli thioredoxin reductase refined at 2 A resolution. Implications for a large conformational change during catalysis.
J.Mol.Biol., 236, 1994
|
|
1SPR
 
 | |
1TDE
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI THIOREDOXIN REDUCTASE REFINED AT 2 ANGSTROM RESOLUTION: IMPLICATIONS FOR A LARGE CONFORMATIONAL CHANGE DURING CATALYSIS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, THIOREDOXIN REDUCTASE | | Authors: | Waksman, G, Krishna, T.S.R, Williams Junior, C.H, Kuriyan, J. | | Deposit date: | 1994-01-14 | | Release date: | 1994-11-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Escherichia coli thioredoxin reductase refined at 2 A resolution. Implications for a large conformational change during catalysis.
J.Mol.Biol., 236, 1994
|
|
2BHM
 
 | | Crystal structure of VirB8 from Brucella suis | | Descriptor: | TYPE IV SECRETION SYSTEM PROTEIN VIRB8 | | Authors: | Bayliss, R, Baron, C, Waksman, G. | | Deposit date: | 2005-01-14 | | Release date: | 2005-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Two Core Subunits of the Bacterial Type Iv Secretion System, Virb8 from Brucella Suis and Comb10 from Helicobacter Pylori
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2THF
 
 | | STRUCTURE OF HUMAN ALPHA-THROMBIN Y225F MUTANT BOUND TO D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, THROMBIN HEAVY CHAIN, ... | | Authors: | Caccia, S, Futterer, K, Di Cera, E, Waksman, G. | | Deposit date: | 1999-01-26 | | Release date: | 1999-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected crucial role of residue 225 in serine proteases.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1EKB
 
 | | THE SERINE PROTEASE DOMAIN OF ENTEROPEPTIDASE BOUND TO INHIBITOR VAL-ASP-ASP-ASP-ASP-LYS-CHLOROMETHANE | | Descriptor: | ENTEROPEPTIDASE, VAL-ASP-ASP-ASP-ASP-LYK PEPTIDE, ZINC ION | | Authors: | Fuetterer, K, Lu, D, Sadler, J.E, Waksman, G. | | Deposit date: | 1999-05-02 | | Release date: | 1999-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of enteropeptidase light chain complexed with an analog of the trypsinogen activation peptide.
J.Mol.Biol., 292, 1999
|
|
3CL3
 
 | | Crystal Structure of a vFLIP-IKKgamma complex: Insights into viral activation of the IKK signalosome | | Descriptor: | NF-kappa-B essential modulator, ORF K13 | | Authors: | Bagneris, C, Ageichik, A.V, Cronin, N, Boshoff, C, Waksman, G, Barrett, T. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a vFlip-IKKgamma complex: insights into viral activation of the IKK signalosome.
Mol.Cell, 30, 2008
|
|
3CRE
 
 | |
1NLY
 
 | | Crystal structure of the traffic ATPase of the Helicobacter pylori type IV secretion system in complex with ATPgammaS | | Descriptor: | MAGNESIUM ION, NONAETHYLENE GLYCOL, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Savvides, S.N, Yeo, H.J, Beck, M.R, Blaesing, F, Lurz, R, Lanka, E, Buhrdorf, R, Fischer, W, Haas, R, Waksman, G. | | Deposit date: | 2003-01-08 | | Release date: | 2003-05-06 | | Last modified: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | VirB11 ATPases are dynamic hexameric assemblies: New insights into bacterial type IV secretion
Embo J., 22, 2003
|
|
1NZV
 
 | |
5KTQ
 
 | | LARGE FRAGMENT OF TAQ DNA POLYMERASE BOUND TO DCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, PROTEIN (DNA POLYMERASE I) | | Authors: | Li, Y, Kong, Y, Korolev, S, Waksman, G. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates.
Protein Sci., 7, 1998
|
|
6Y7S
 
 | | 2.85 A cryo-EM structure of the in vivo assembled type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Zyla, D, Hospenthal, M, Waksman, G, Glockshuber, R. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-31 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
5CDW
 
 | | Crystal Structure Analysis of a mutant Grb2 SH2 domain (W121G) with a pYVNV peptide | | Descriptor: | Growth factor receptor-bound protein 2, SER-PTR-VAL-ASN-VAL-GLN | | Authors: | Papaioannou, D, Geibel, S, Kunze, M, Kay, C, Waksman, G. | | Deposit date: | 2015-07-05 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structural and biophysical investigation of the interaction of a mutant Grb2 SH2 domain (W121G) with its cognate phosphopeptide.
Protein Sci., 25, 2016
|
|
5FLU
 
 | | Structure of a Chaperone-Usher pilus reveals the molecular basis of rod uncoilin | | Descriptor: | PAP FIMBRIAL MAJOR PILIN PROTEIN | | Authors: | Hospenthal, M.K, Redzej, A, Dodson, K, Ukleja, M, Frenz, B, Hultgren, S.J, DiMaio, F, Egelman, E.H, Waksman, G. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a Chaperone-Usher Pilus Reveals the Molecular Basis of Rod Uncoiling.
Cell(Cambridge,Mass.), 164, 2016
|
|
6QCM
 
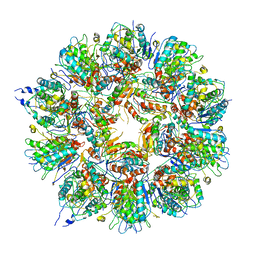 | | Cryo em structure of the Listeria stressosome | | Descriptor: | RsbR protein, RsbR protein,RsbR protein, RsbS protein | | Authors: | Williams, A.H, Redzej, A, Waksman, G, Cossart, P. | | Deposit date: | 2018-12-28 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | The cryo-electron microscopy supramolecular structure of the bacterial stressosome unveils its mechanism of activation.
Nat Commun, 10, 2019
|
|
4L0J
 
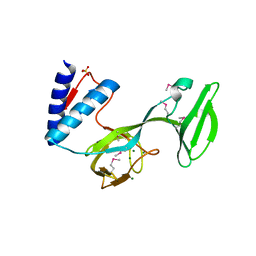 | | Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems | | Descriptor: | DNA helicase I, MAGNESIUM ION, SULFATE ION | | Authors: | Redzej, A, Ilangovan, A, Lang, S, Gruber, C.J, Topf, M, Zangger, K, Zechner, E.L, Waksman, G. | | Deposit date: | 2013-05-31 | | Release date: | 2013-06-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems.
Mol.Microbiol., 89, 2013
|
|
5O96
 
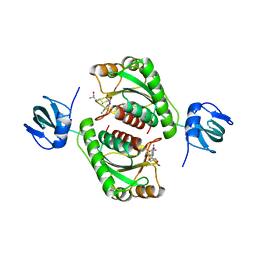 | |
5O95
 
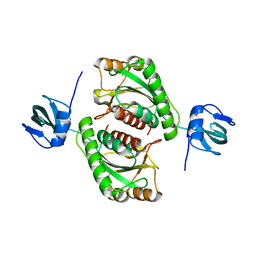 | |
5OH0
 
 | | The Cryo-Electron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Hospenthal, M.K, Costa, T.R.D, Redzej, A, Waksman, G. | | Deposit date: | 2017-07-13 | | Release date: | 2017-11-22 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The Cryoelectron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod.
Structure, 25, 2017
|
|
2YPW
 
 | | Atomic model for the N-terminus of TraO fitted in the full-length structure of the bacterial pKM101 type IV secretion system core complex | | Descriptor: | TRAO | | Authors: | Rivera-Calzada, A, Fronzes, R, Savva, C.G, Chandran, V, Lian, P.W, Laeremans, T, Pardon, E, Steyaert, J, Remaut, H, Waksman, G, Orlova, E.V. | | Deposit date: | 2012-11-02 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.4 Å) | | Cite: | Structure of a Bacterial Type Iv Secretion Core Complex at Subnanometre Resolution.
Embo J., 32, 2013
|
|
3ZBI
 
 | | Fitting result in the O-layer of the subnanometer structure of the bacterial pKM101 type IV secretion system core complex digested with elastase | | Descriptor: | TRAF PROTEIN, TRAN PROTEIN, TRAO PROTEIN | | Authors: | Rivera-Calzada, A, Fronzes, R, Savva, C.G, Chandran, V, Lian, P.W, Laeremans, T, Pardon, E, Steyaert, J, Remaut, H, Waksman, G, Orlova, E.V. | | Deposit date: | 2012-11-10 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structure of a Bacterial Type Iv Secretion Core Complex at Subnanometre Resolution.
Embo J., 32, 2013
|
|
2BHV
 
 | | Structure of ComB10 of the Com Type IV secretion system of Helicobacter pylori | | Descriptor: | COMB10 | | Authors: | Terradot, L, Oomen, C, Bayliss, R, Leonard, G, Waksman, G. | | Deposit date: | 2005-01-18 | | Release date: | 2005-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of Two Core Subunits of the Bacterial Type Iv Secretion System, Virb8 from Brucella Suis and Comb10 from Helicobacter Pylori
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
