1WLF
 
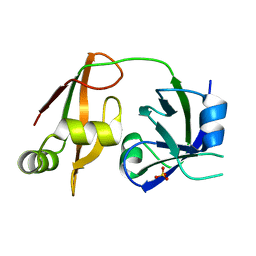 | | Structure of the N-terminal domain of PEX1 AAA-ATPase: Characterization of a putative adaptor-binding domain | | Descriptor: | Peroxisome biogenesis factor 1, SULFATE ION | | Authors: | Shiozawa, K, Maita, N, Tomii, K, Seto, A, Goda, N, Tochio, H, Akiyama, Y, Shimizu, T, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2004-06-25 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the N-terminal Domain of PEX1 AAA-ATPase: CHARACTERIZATION OF A PUTATIVE ADAPTOR-BINDING DOMAIN
J.Biol.Chem., 279, 2004
|
|
3B44
 
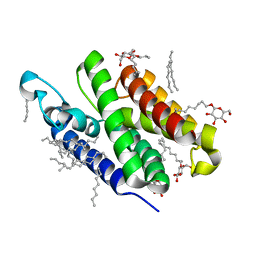 | | Crystal structure of GlpG W136A mutant | | Descriptor: | glpG, nonyl beta-D-glucopyranoside | | Authors: | Wang, Y, Maegawa, S, Akiyama, Y, Ha, Y. | | Deposit date: | 2007-10-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG.
J.Mol.Biol., 374, 2007
|
|
3B45
 
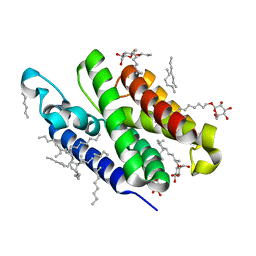 | | Crystal structure of GlpG at 1.9A resolution | | Descriptor: | glpG, nonyl beta-D-glucopyranoside | | Authors: | Wang, Y, Maegawa, S, Akiyama, Y, Ha, Y. | | Deposit date: | 2007-10-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG.
J.Mol.Biol., 374, 2007
|
|
6A8V
 
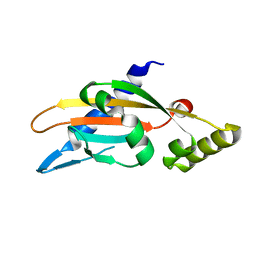 | | PhoQ sensor domain (D179R mutant): analysis of internal cavity | | Descriptor: | Sensor protein PhoQ | | Authors: | Yoshitani, K, Ishii, E, Taniguchi, K, Sugimoto, H, Shiro, Y, Mori, H, Akiyama, Y, Kato, A, Utsumi, R, Eguchi, Y. | | Deposit date: | 2018-07-10 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of an internal cavity in the PhoQ sensor domain for PhoQ activity and SafA-mediated control.
Biosci. Biotechnol. Biochem., 83, 2019
|
|
6A8U
 
 | | PhoQ sensor domain (wild type): analysis of internal cavity | | Descriptor: | Sensor protein PhoQ | | Authors: | Yoshitani, K, Ishii, E, Taniguchi, K, Sugimoto, H, Shiro, Y, Mori, H, Akiyama, Y, Kato, A, Utsumi, R, Eguchi, Y. | | Deposit date: | 2018-07-10 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Identification of an internal cavity in the PhoQ sensor domain for PhoQ activity and SafA-mediated control.
Biosci. Biotechnol. Biochem., 83, 2019
|
|
8WSM
 
 | | NLRP3 NACHT domain in complex with compound 32 | | Descriptor: | 2-[[2-methyl-5-(trifluoromethyl)phenyl]amino]-~{N}-(1,4-oxazepan-4-ylsulfonyl)-1,3-oxazole-4-carboxamide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Akai, S, Orita, T, Adachi, T. | | Deposit date: | 2023-10-17 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Novel NLRP3 Inflammasome Inhibitors Composed of an Oxazole Scaffold Bearing an Acylsulfamide.
Acs Med.Chem.Lett., 14, 2023
|
|
5B1S
 
 | | Crystal structure of Trypanosoma cruzi spermidine synthase in complex with 2-(2-fluorophenyl)ethanamine | | Descriptor: | 2-(2-fluorophenyl)ethanamine, 5'-[(S)-(3-AMINOPROPYL)(METHYL)-LAMBDA~4~-SULFANYL]-5'-DEOXYADENOSINE, Spermidine synthase, ... | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | In silico, in vitro, X-ray crystallography, and integrated strategies for discovering spermidine synthase inhibitors for Chagas disease
Sci Rep, 7, 2017
|
|
7W6Y
 
 | | Crystal structure of Kangiella koreensis RseP orthologue in complex with batimastat in space group P1 | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, Anti sigma-E protein, RseA, ... | | Authors: | Imaizumi, Y, Takanuki, K, Nogi, T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanistic insights into intramembrane proteolysis by E. coli site-2 protease homolog RseP.
Sci Adv, 8, 2022
|
|
7W6X
 
 | |
7W6Z
 
 | |
7W70
 
 | |
7W71
 
 | |
2ZPL
 
 | | Crystal structure analysis of PDZ domain A | | Descriptor: | GLYCEROL, NICKEL (II) ION, Regulator of sigma E protease | | Authors: | Inaba, K, Suzuki, M. | | Deposit date: | 2008-07-17 | | Release date: | 2008-10-21 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure analysis of PDZ-domain A
To be Published
|
|
2ZPM
 
 | |
3UBB
 
 | |
5AZV
 
 | | Crystal structure of hPPARgamma ligand binding domain complexed with 17-oxoDHA | | Descriptor: | (4~{Z},7~{Z},10~{Z},13~{Z},19~{Z})-17-oxidanylidenedocosa-4,7,10,13,19-pentaenoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2015-10-23 | | Release date: | 2016-07-06 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 17-OxoDHA Is a PPAR alpha/gamma Dual Covalent Modifier and Agonist
Acs Chem.Biol., 2016
|
|
5AZT
 
 | | Ternary complex of hPPARalpha ligand binding domain, 17-oxoDHA and a SRC1 peptide | | Descriptor: | (4~{Z},7~{Z},10~{Z},13~{Z},19~{Z})-17-oxidanylidenedocosa-4,7,10,13,19-pentaenoic acid, 15-meric peptide from Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor alpha | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2015-10-23 | | Release date: | 2016-07-06 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | 17-OxoDHA Is a PPAR alpha/gamma Dual Covalent Modifier and Agonist
Acs Chem.Biol., 2016
|
|
7WI3
 
 | | Cryo-EM structure of E.Coli FtsH-HflkC AAA protease complex | | Descriptor: | ATP-dependent zinc metalloprotease FtsH, Modulator of FtsH protease HflC, Modulator of FtsH protease HflK | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2022-01-02 | | Release date: | 2022-06-01 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the entire FtsH-HflKC AAA protease complex.
Cell Rep, 39, 2022
|
|
7WI4
 
 | | Cryo-EM structure of E.Coli FtsH protease cytosolic domains | | Descriptor: | ATP-dependent zinc metalloprotease FtsH, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2022-01-02 | | Release date: | 2022-06-01 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the entire FtsH-HflKC AAA protease complex.
Cell Rep, 39, 2022
|
|
5Y4Q
 
 | | Crystal structure of Trypanosoma cruzi spermidine synthase in complex with N-(4-methoxyphenyl)quinolin-4-amine | | Descriptor: | 5'-[(S)-(3-AMINOPROPYL)(METHYL)-LAMBDA~4~-SULFANYL]-5'-DEOXYADENOSINE, N-(4-methoxyphenyl)quinolin-4-amine, Spermidine synthase, ... | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of a Hidden Trypanosoma cruzi Spermidine Synthase Binding Site and Inhibitors through In Silico, In Vitro , and X-ray Crystallography.
Acs Omega, 8, 2023
|
|
5Y4P
 
 | | Crystal structure of Trypanosoma cruzi spermidine synthase in complex with 5-methoxy-2-(5-methyl-4,5-dihydro-1H-imidazol-2-yl)phenol | | Descriptor: | 5'-[(S)-(3-AMINOPROPYL)(METHYL)-LAMBDA~4~-SULFANYL]-5'-DEOXYADENOSINE, 5-methoxy-2-[(5R)-5-methyl-4,5-dihydro-1H-imidazol-2-yl]phenol, Spermidine synthase, ... | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of a Hidden Trypanosoma cruzi Spermidine Synthase Binding Site and Inhibitors through In Silico, In Vitro , and X-ray Crystallography.
Acs Omega, 8, 2023
|
|
6AIT
 
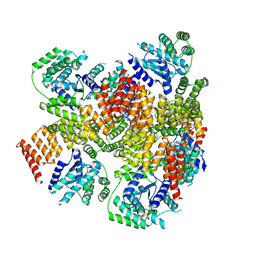 | | Crystal structure of E. coli BepA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-barrel assembly-enhancing protease, ZINC ION | | Authors: | Umar, M.S.M, Tanaka, Y, Kamikubo, H, Tsukazaki, T. | | Deposit date: | 2018-08-24 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural Basis for the Function of the beta-Barrel Assembly-Enhancing Protease BepA.
J. Mol. Biol., 431, 2019
|
|
3WKL
 
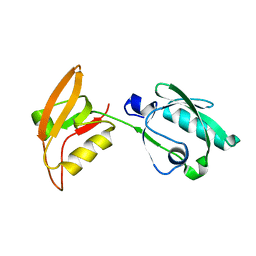 | |
3WKM
 
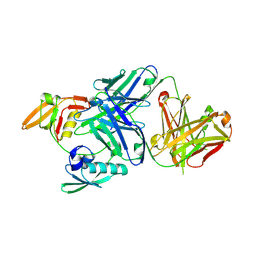 | | The periplasmic PDZ tandem fragment of the RseP homologue from Aquifex aeolicus in complex with the Fab fragment | | Descriptor: | MOUSE IGG1-KAPPA FAB (HEAVY CHAIN), MOUSE IGG1-KAPPA FAB (LIGHT CHAIN), Putative zinc metalloprotease aq_1964 | | Authors: | Nogi, T, Tabata, S, Tamura-kawakami, K, Takagi, J. | | Deposit date: | 2013-10-28 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Structure-Based Model of Substrate Discrimination by a Noncanonical PDZ Tandem in the Intramembrane-Cleaving Protease RseP
Structure, 22, 2013
|
|
5XI8
 
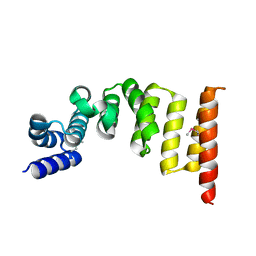 | | Structure and function of the TPR domain | | Descriptor: | Beta-barrel assembly-enhancing protease | | Authors: | Tanaka, Y, Tsukazaki, T. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The TPR domain of BepA is required for productive interaction with substrate proteins and the beta-barrel assembly machinery complex.
Mol. Microbiol., 106, 2017
|
|
