1WYW
 
 | | Crystal Structure of SUMO1-conjugated thymine DNA glycosylase | | Descriptor: | CHLORIDE ION, G/T mismatch-specific thymine DNA glycosylase, MAGNESIUM ION, ... | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-02-17 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thymine DNA glycosylase conjugated to SUMO-1.
Nature, 435, 2005
|
|
2ASQ
 
 | | Solution Structure of SUMO-1 in Complex with a SUMO-binding Motif (SBM) | | Descriptor: | Protein inhibitor of activated STAT2, Small ubiquitin-related modifier 1 | | Authors: | Song, J, Zhang, Z, Hu, W, Chen, Y. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Small Ubiquitin-like Modifier (SUMO) Recognition of a SUMO Binding Motif: A reversal of the bound orientation
J.Biol.Chem., 280, 2005
|
|
7AAV
 
 | | Human pre-Bact-2 spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, D-chiro inositol hexakisphosphate, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7AOA
 
 | | Structure of the extended MTA1/HDAC1/MBD2/RBBP4 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, Histone-binding protein RBBP4, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (19.4 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
2PE6
 
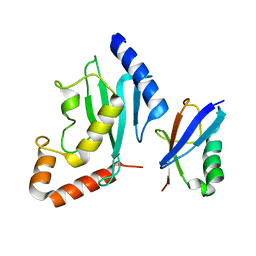 | | Non-covalent complex between human SUMO-1 and human Ubc9 | | Descriptor: | SUMO-conjugating enzyme UBC9, Small ubiquitin-related modifier 1 | | Authors: | Capili, A.D, Lima, C.D. | | Deposit date: | 2007-04-02 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Analysis of a Complex between SUMO and Ubc9 Illustrates Features of a Conserved E2-Ubl Interaction.
J.Mol.Biol., 369, 2007
|
|
1EJ9
 
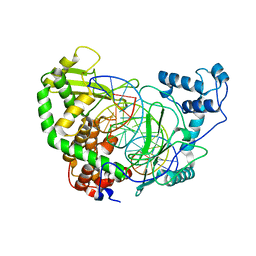 | | CRYSTAL STRUCTURE OF HUMAN TOPOISOMERASE I DNA COMPLEX | | Descriptor: | DNA (5'-D(*C*AP*AP*AP*AP*AP*GP*AP*CP*TP*CP*AP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3'), DNA (5'-D(*C*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*TP*GP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3'), DNA TOPOISOMERASE I | | Authors: | Redinbo, M.R, Champoux, J.J, Hol, W.G. | | Deposit date: | 2000-03-01 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel insights into catalytic mechanism from a crystal structure of human topoisomerase I in complex with DNA.
Biochemistry, 39, 2000
|
|
2VXD
 
 | |
7D58
 
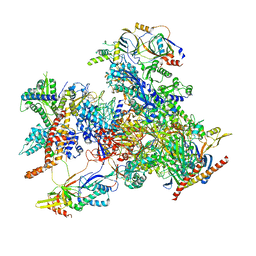 | | cryo-EM structure of human RNA polymerase III in elongating state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D59
 
 | | cryo-EM structure of human RNA polymerase III in apo state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6XOG
 
 | | Structure of SUMO1-ML786519 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOH
 
 | | Structure of SUMO1-ML00789344 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOI
 
 | | Structure of SUMO1-ML00752641 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6ID0
 
 | | Cryo-EM structure of a human intron lariat spliceosome prior to Prp43 loaded (ILS1 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6Z2J
 
 | | The structure of the dimeric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
6ID1
 
 | | Cryo-EM structure of a human intron lariat spliceosome after Prp43 loaded (ILS2 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6Z2K
 
 | | The structure of the tetrameric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
8X1C
 
 | | Structure of nucleosome-bound SRCAP-C in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of nucleosome-bound SRCAP-C in the ADP-bound state
To Be Published
|
|
8X19
 
 | | Structure of nucleosome-bound SRCAP-C in the ADP-BeFx-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of nucleosome-bound SRCAP-C in the ADP-BeFx-bound state
To Be Published
|
|
8Y6O
 
 | | Cryo-EM Structure of the human minor pre-B complex (pre-precatalytic spliceosome) U11 and tri-snRNP part | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Centrosomal AT-AC splicing factor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bai, R, Yuan, M, Zhang, P, Luo, T, Shi, Y, Wan, R. | | Deposit date: | 2024-02-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome.
Science, 383, 2024
|
|
8ITY
 
 | | human RNA polymerase III pre-initiation complex closed DNA 1 | | Descriptor: | DNA (82-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Hou, H, Jin, Q, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the SNAPc-bound RNA polymerase III preinitiation complex.
Cell Res., 33, 2023
|
|
8A0B
 
 | | Inhibitor binding to HDAC2 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dihydroisoindol-2-yl-[(2R,4S)-4-phenylpyrrolidin-1-ium-2-yl]methanone, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-27 | | Release date: | 2022-09-21 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZS
 
 | | HDAC2 complexed with an inhibitory ligand | | Descriptor: | (5~{S})-5-(4-chlorophenyl)pyrrolidin-2-one, 1,2-ETHANEDIOL, 2-(cyclohexylazaniumyl)ethanesulfonate, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZP
 
 | | Structure of HDAC2 complexed with an inhibitory ligand | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZR
 
 | | HDAC2 in complex with inhibitory ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZO
 
 | | HDAC2 in complex with an inhibitor | | Descriptor: | 2-(cyclohexylazaniumyl)ethanesulfonate, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-21 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
