7K17
 
 | |
3W03
 
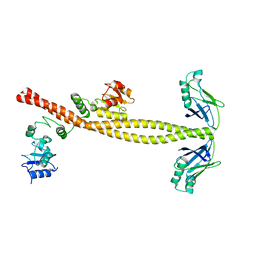
 | | XLF-XRCC4 complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Wu, Q, Ochi, T, Matak-Vinkovic, D, Robinson, C.V, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2012-10-17 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (8.492 Å) | | Cite: | Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions
Biochem.Soc.Trans., 39, 2011
|
|
3II6
 
 | |
1IK9
 
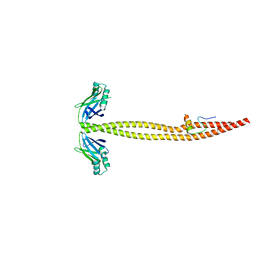 | | CRYSTAL STRUCTURE OF A XRCC4-DNA LIGASE IV COMPLEX | | Descriptor: | DNA LIGASE IV, DNA REPAIR PROTEIN XRCC4 | | Authors: | Sibanda, B.L, Critchlow, S.E, Begun, J, Pei, X.Y, Jackson, S.P, Blundell, T.L, Pellegrini, L. | | Deposit date: | 2001-05-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an Xrcc4-DNA ligase IV complex.
Nat.Struct.Biol., 8, 2001
|
|
7ZWA
 
 | | CryoEM structure of Ku heterodimer bound to DNA and PAXX | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
7ZYG
 
 | | CryoEM structure of Ku heterodimer bound to DNA, PAXX and XLF | | Descriptor: | DNA, Non-homologous end-joining factor 1, PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
1RW2
 
 | | Three-dimensional structure of Ku80 CTD | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Zhang, Z, Hu, W, Cano, L, Lee, T.D, Chen, D.J, Chen, Y. | | Deposit date: | 2003-12-15 | | Release date: | 2003-12-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions.
STRUCTURE, 12, 2004
|
|
6ABO
 
 | | human XRCC4 and IFFO1 complex | | Descriptor: | DNA repair protein XRCC4, GLYCEROL, Intermediate filament family orphan 1, ... | | Authors: | Li, J, Liu, L, Liang, H, Liu, Y, Xu, D. | | Deposit date: | 2018-07-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The nucleoskeleton protein IFFO1 immobilizes broken DNA and suppresses chromosome translocation during tumorigenesis.
Nat.Cell Biol., 21, 2019
|
|
3Q4F
 
 | | Crystal structure of xrcc4/xlf-cernunnos complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Ropars, V, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2010-12-23 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3KGV
 
 | |
8X2R
 
 | | The Crystal Structure of HSP 90-alpha from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2023-11-10 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of HSP 90-alpha from Biortus.
To Be Published
|
|
3U3Z
 
 | | Structure of human microcephalin (MCPH1) tandem BRCT domains in complex with an H2A.X peptide phosphorylated at Ser139 and Tyr142 | | Descriptor: | GLYCEROL, Histone H2A.X peptide, Microcephalin | | Authors: | Singh, N, Thompson, J.R, Heroux, A, Mer, G. | | Deposit date: | 2011-10-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dual recognition of phosphoserine and phosphotyrosine in histone variant H2A.X by DNA damage response protein MCPH1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8ASC
 
 | | Ku70/80 binds to the Ku-binding motif of PAXX | | Descriptor: | DNA (5'-D(P*CP*GP*GP*AP*TP*CP*GP*AP*GP*GP*GP*CP*CP*CP*GP*AP*TP*AP*T)-3'), DNA (5'-D(P*GP*GP*GP*CP*CP*CP*TP*CP*GP*AP*TP*CP*CP*G)-3'), Protein PAXX, ... | | Authors: | Seif El Dahan, M, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
4U93
 
 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7R)-2-amino-7-[4-fluoro-2-(6-methoxypyridin-2-yl)phenyl]-4-methyl-7,8-dihydropyrido[4,3-d]pyrimidin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-05 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
4OQA
 
 | | Structure of Human PARP-1 bound to a DNA double strand break in complex with (2Z)-2-(2,4-dihydroxybenzylidene)-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide | | Descriptor: | (2Z)-2-(2,4-dihydroxybenzylidene)-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide, DNA (26-MER), Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Steffen, J.D. | | Deposit date: | 2014-02-07 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Discovery and Structure-Activity Relationship of Novel 2,3-Dihydrobenzofuran-7-carboxamide and 2,3-Dihydrobenzofuran-3(2H)-one-7-carboxamide Derivatives as Poly(ADP-ribose)polymerase-1 Inhibitors.
J.Med.Chem., 57, 2014
|
|
2QM4
 
 | | Crystal structure of human XLF/Cernunnos, a non-homologous end-joining factor | | Descriptor: | Non-homologous end-joining factor 1 | | Authors: | Li, Y, Chirgadze, D.Y, Sibanda, B.L, Bolanos-Garcia, V.M, Davies, O.R, Blundell, T.L. | | Deposit date: | 2007-07-14 | | Release date: | 2007-12-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ.
Embo J., 27, 2008
|
|
4DQY
 
 | |
5UPP
 
 | | Crystal structure of human fumarate hydratase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fumarate hydratase, mitochondrial | | Authors: | Rangel, V.L, Ajalla, M.A, Rustiguel, J.K, Pereira de Padua, R.A, Nonato, M.C. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, biochemical and biophysical characterization of recombinant human fumarate hydratase.
Febs J., 2019
|
|
6CYG
 
 | | Hsp90-alpha N-domain bound to NEOCA | | Descriptor: | 5'-N-(2-HYDROXYL)ETHYL CARBOXYAMIDO ADENOSINE, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Huck, J.D, Campomizzi, C, Gewirth, D.T. | | Deposit date: | 2018-04-05 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.503922 Å) | | Cite: | NECA derivatives exploit the paralog-specific properties of the site 3 side pocket of Grp94, the endoplasmic reticulum Hsp90.
J.Biol.Chem., 294, 2019
|
|
6CYH
 
 | | Hsp90-alpha N-domain bound to NEACA | | Descriptor: | 5'-N-[(2-AMINO)ETHYL CARBOXAMIDO] ADENOSINE, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Huck, J.D, Campomizzi, C, Gewirth, D.T. | | Deposit date: | 2018-04-05 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | NECA derivatives exploit the paralog-specific properties of the site 3 side pocket of Grp94, the endoplasmic reticulum Hsp90.
J.Biol.Chem., 294, 2019
|
|
7LUB
 
 | |
4OPX
 
 | | Structure of Human PARP-1 bound to a DNA double strand break in complex with (2R)-5-fluoro-2-methyl-2,3-dihydro-1-benzofuran-7-carboxamide | | Descriptor: | (2R)-5-fluoro-2-methyl-2,3-dihydro-1-benzofuran-7-carboxamide, DNA (26-MER), Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Steffen, J.D. | | Deposit date: | 2014-02-06 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.314 Å) | | Cite: | Discovery and Structure-Activity Relationship of Novel 2,3-Dihydrobenzofuran-7-carboxamide and 2,3-Dihydrobenzofuran-3(2H)-one-7-carboxamide Derivatives as Poly(ADP-ribose)polymerase-1 Inhibitors.
J.Med.Chem., 57, 2014
|
|
4OQB
 
 | | Structure of Human PARP-1 bound to a DNA double strand break in complex with (2Z)-2-{4-[2-(morpholin-4-yl)ethoxy]benzylidene}-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide | | Descriptor: | (2Z)-2-{4-[2-(morpholin-4-yl)ethoxy]benzylidene}-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide, DNA (26-MER), Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Steffen, J.D. | | Deposit date: | 2014-02-07 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.362 Å) | | Cite: | Discovery and Structure-Activity Relationship of Novel 2,3-Dihydrobenzofuran-7-carboxamide and 2,3-Dihydrobenzofuran-3(2H)-one-7-carboxamide Derivatives as Poly(ADP-ribose)polymerase-1 Inhibitors.
J.Med.Chem., 57, 2014
|
|
3HHU
 
 | | Human heat-shock protein 90 (HSP90) in complex with {4-[3-(2,4-dihydroxy-5-isopropyl-phenyl)-5-thioxo- 1,5-dihydro-[1,2,4]triazol-4-yl]-benzyl}-carbamic acid ethyl ester {ZK 2819} | | Descriptor: | Heat shock protein HSP 90-alpha, ethyl (4-{3-[2,4-dihydroxy-5-(1-methylethyl)phenyl]-5-sulfanyl-4H-1,2,4-triazol-4-yl}benzyl)carbamate | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2009-05-17 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Potent triazolothione inhibitor of heat-shock protein-90.
Chem.Biol.Drug Des., 74, 2009
|
|
3WQ9
 
 | | Crystal structure of Hsp90-alpha N-terminal domain in complex with 2-(4-Hydroxy-cyclohexylamino)-4-[5-(4-phenyl-imidazol-1-yl)-isoquinolin-1-yl]-benzamide | | Descriptor: | 2-[(trans-4-hydroxycyclohexyl)amino]-4-[5-(4-phenyl-1H-imidazol-1-yl)isoquinolin-1-yl]benzamide, Heat shock protein HSP 90-alpha | | Authors: | Chong, K.T, Yamashita, S, Oshiumi, H, Uno, T, Kitade, M. | | Deposit date: | 2014-01-23 | | Release date: | 2015-02-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of highly selective Hsp90 / inhibitors by structure and thermodynamics guided design
To be Published
|
|
