1F9E
 
 | | CASPASE-8 SPECIFICITY PROBED AT SUBSITE S4: CRYSTAL STRUCTURE OF THE CASPASE-8-Z-DEVD-CHO | | Descriptor: | (PHQ)DEVD, CASPASE-8 ALPHA CHAIN, CASPASE-8 BETA CHAIN | | Authors: | Blanchard, H, Donepudi, M, Tschopp, M, Kodandapani, L, Wu, J.C, Grutter, M.G. | | Deposit date: | 2000-07-10 | | Release date: | 2001-07-10 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Caspase-8 specificity probed at subsite S(4): crystal structure of the caspase-8-Z-DEVD-cho complex.
J.Mol.Biol., 302, 2000
|
|
1IBC
 
 | |
1GFW
 
 | |
1ICE
 
 | | STRUCTURE AND MECHANISM OF INTERLEUKIN-1BETA CONVERTING ENZYME | | Descriptor: | INTERLEUKIN-1 BETA CONVERTING ENZYME, TETRAPEPTIDE ALDEHYDE | | Authors: | Wilson, K.P, Griffith, J.P, Kim, E.E, Navia, M.A. | | Deposit date: | 1994-09-29 | | Release date: | 1995-07-28 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of interleukin-1 beta converting enzyme.
Nature, 370, 1994
|
|
3EDQ
 
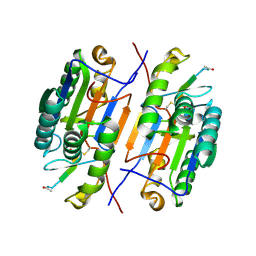 | | Crystal structure of Caspase-3 with inhibitor AC-LDESD-CHO | | Descriptor: | AC-LDESD-CHO peptide, Caspase-3 | | Authors: | Fu, G. | | Deposit date: | 2008-09-03 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural basis for executioner caspase recognition of P5 position in substrates.
Apoptosis, 13, 2008
|
|
3H13
 
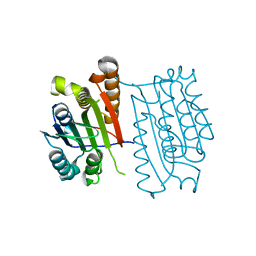 | | c-FLIPL protease-like domain | | Descriptor: | CASP8 and FADD-like apoptosis regulator | | Authors: | Jeffrey, P.D, Yu, J.W, Shi, Y. | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of procaspase-8 activation by c-FLIPL.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GJS
 
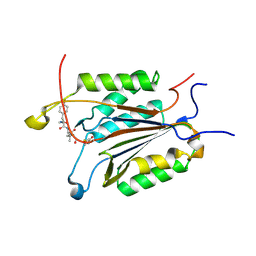 | | Caspase-3 Binds Diverse P4 Residues in Peptides | | Descriptor: | Ac-YVAD-Cho inhibitor, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Fang, B, Fu, G, Agniswamy, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2009-03-09 | | Release date: | 2009-03-24 | | Last modified: | 2011-08-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Apoptosis, 14, 2009
|
|
3H1P
 
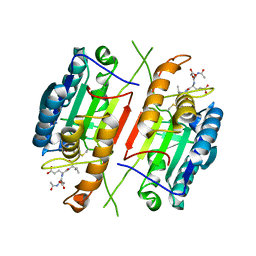 | |
3E4C
 
 | | Procaspase-1 zymogen domain crystal structure | | Descriptor: | Caspase-1, MAGNESIUM ION | | Authors: | Elliott, J.M, Rouge, L, Wiesmann, C, Scheer, J.M. | | Deposit date: | 2008-08-11 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of procaspase-1 zymogen domain reveals insight into inflammatory caspase autoactivation
J.Biol.Chem., 284, 2009
|
|
3EDR
 
 | |
3H11
 
 | |
3GJR
 
 | | Caspase-3 Binds Diverse P4 Residues in Peptides | | Descriptor: | Caspase-3 subunit p12, Caspase-3 subunit p17, GLYCEROL, ... | | Authors: | Fang, B, Fu, G, Agniswamy, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2009-03-09 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Apoptosis, 14, 2009
|
|
3GJT
 
 | | Caspase-3 Binds Diverse P4 Residues in Peptides | | Descriptor: | Caspase-3 subunit p12, Caspase-3 subunit p17, peptide inhibitor | | Authors: | Fang, B, Fu, G, Agniswamy, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2009-03-09 | | Release date: | 2009-03-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Apoptosis, 14, 2009
|
|
3H0E
 
 | | 3,4-Dihydropyrimido(1,2-a)indol-10(2H)-ones as Potent Non-Peptidic Inhibitors of Caspase-3 | | Descriptor: | (10S)-3,3-dimethyl-8-{[(2S)-2-(phenoxymethyl)pyrrolidin-1-yl]sulfonyl}-2,3,4,10-tetrahydropyrimido[1,2-a]indol-10-ol, Caspase-3 | | Authors: | Xu, W. | | Deposit date: | 2009-04-09 | | Release date: | 2009-11-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | 3,4-Dihydropyrimido(1,2-a)indol-10(2H)-ones as potent non-peptidic inhibitors of caspase-3
Bioorg.Med.Chem., 17, 2009
|
|
3GJQ
 
 | | Caspase-3 Binds Diverse P4 Residues in Peptides | | Descriptor: | Caspase-3 subunit p12, Caspase-3 subunit p17, peptide inhibitor | | Authors: | Fang, B, Fu, G, Agniswamy, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2009-03-09 | | Release date: | 2009-03-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Apoptosis, 14, 2009
|
|
3IBF
 
 | | Crystal structure of unliganded caspase-7 | | Descriptor: | Caspase-7 | | Authors: | Agniswamy, J. | | Deposit date: | 2009-07-15 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational similarity in the activation of caspase-3 and -7 revealed by the unliganded and inhibited structures of caspase-7.
Apoptosis, 14, 2009
|
|
4M9R
 
 | | Crystal structure of CED-3 | | Descriptor: | Cell death protein 3 | | Authors: | Xu, Y, Jeffrey, P.D, Shi, Y.G. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.656 Å) | | Cite: | Mechanistic insights into CED-4-mediated activation of CED-3
Genes Dev., 27, 2013
|
|
7QP1
 
 | | Crystal structure of metacaspase from candida glabrata with calcium | | Descriptor: | CALCIUM ION, CHLORIDE ION, Metacaspase-1 | | Authors: | Conchou, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and molecular determinants of Candida glabrata metacaspase maturation and activation by calcium.
Commun Biol, 5, 2022
|
|
7QP0
 
 | | Crystal structure of metacaspase from candida glabrata with magnesium | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, Metacaspase-1 | | Authors: | Conchou, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and molecular determinants of Candida glabrata metacaspase maturation and activation by calcium.
Commun Biol, 5, 2022
|
|
3S70
 
 | | Crystal structure of active caspase-6 bound with Ac-VEID-CHO solved by As-SAD | | Descriptor: | ACETATE ION, CACODYLATE ION, Caspase-6, ... | | Authors: | Su, X.-D, Liu, X, Wang, X.-J. | | Deposit date: | 2011-05-26 | | Release date: | 2012-04-11 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.625 Å) | | Cite: | Get phases from arsenic anomalous scattering: de novo SAD phasing of two protein structures crystallized in cacodylate buffer
Plos One, 6, 2011
|
|
4NBN
 
 | |
4N5D
 
 | | Tailoring Small Molecules for an Allosteric Site on Procaspase-6: Cpd1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-amino-2,8-dimethylpyrido[2,3-d]pyrimidin-7(8H)-one, Caspase-6, ... | | Authors: | Murray, J.M, Steffek, M. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Tailoring small molecules for an allosteric site on procaspase-6.
Chemmedchem, 9, 2014
|
|
8DGZ
 
 | | Caspase-7 bound to substrate mimic and allosteric inhibitor | | Descriptor: | 2-{[2-(4-chlorophenyl)-2-oxoethyl]sulfanyl}benzoic acid, Ac-Asp-Glu-Val-Asp-Aldehyde, Caspase-7 | | Authors: | Propp, J, Kalenkiewicz, A, Kathryn, F.H, Spies, M.A. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric Tuning of Caspase-7: Establishing the Nexus of Structure and Catalytic Power.
Chemistry, 29, 2023
|
|
8DJ3
 
 | | Caspase-7 bound to novel allosteric inhibitor | | Descriptor: | 2-[(2-{[(3s,5s,7s)-adamantan-1-yl]sulfamoyl}phenyl)sulfanyl]benzoic acid, Caspase-7 | | Authors: | Propp, J, Kalenkiewicz, A, Kathryn, F.H, Spies, M.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Allosteric Tuning of Caspase-7: Establishing the Nexus of Structure and Catalytic Power.
Chemistry, 29, 2023
|
|
7RND
 
 | | Crystal structure of caspase-3 with inhibitor Ac-VDPVD-CHO | | Descriptor: | Ac-VDPVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
