1SAF
 
 | |
8F2I
 
 | | P53 monomer structure | | Descriptor: | Cellular tumor antigen p53 | | Authors: | Solares, M, Kelly, D.F. | | Deposit date: | 2022-11-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-Resolution Imaging of Human Cancer Proteins Using Microprocessor Materials.
Chembiochem, 23, 2022
|
|
8F2H
 
 | |
7BWN
 
 | | Crystal Structure of a Designed Protein Heterocatenane | | Descriptor: | Cellular tumor antigen p53, Chimera of Green fluorescent protein and p53dim | | Authors: | Liu, Y.J, Duan, Z.L, Fang, J, Zhang, F, Xiao, J.Y, Zhang, W.B. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Cellular Synthesis and X-ray Crystal Structure of a Designed Protein Heterocatenane.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5XZC
 
 | | Cryo-EM structure of p300-p53 protein complex | | Descriptor: | Cellular tumor antigen p53, Histone acetyltransferase p300 | | Authors: | Ghosh, R, Roy, S, Sengupta, J. | | Deposit date: | 2017-07-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (10.7 Å) | | Cite: | Tumor suppressor p53-mediated structural reorganization of the transcriptional coactivator p300.
Biochemistry, 2019
|
|
2J10
 
 | | p53 tetramerization domain mutant T329F Q331K | | Descriptor: | CELLULAR TUMOR ANTIGEN P53 | | Authors: | Carbajo, R.J, Mora, P, Sanchez del Pino, M.M, Perez-Paya, E, Pineda-Lucena, A. | | Deposit date: | 2006-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solvent-exposed residues located in the beta-sheet modulate the stability of the tetramerization domain of p53--a structural and combinatorial approach.
Proteins, 71, 2008
|
|
2J11
 
 | | p53 tetramerization domain mutant Y327S T329G Q331G | | Descriptor: | CELLULAR TUMOR ANTIGEN P53 | | Authors: | Carbajo, R.J, Mora, P, Sanchez del Pino, M.M, Perez-Paya, E, Pineda-Lucena, A. | | Deposit date: | 2006-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solvent-exposed residues located in the beta-sheet modulate the stability of the tetramerization domain of p53--a structural and combinatorial approach.
Proteins, 71, 2008
|
|
2J0Z
 
 | | p53 tetramerization domain wild type | | Descriptor: | CELLULAR TUMOR ANTIGEN P53 | | Authors: | Carbajo, R.J, Mora, P, Sanchez del Pino, M.M, Perez-Paya, E, Pineda-Lucena, A. | | Deposit date: | 2006-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solvent-exposed residues located in the beta-sheet modulate the stability of the tetramerization domain of p53--a structural and combinatorial approach.
Proteins, 71, 2008
|
|
4MZR
 
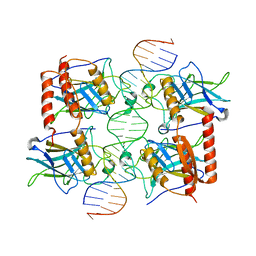 | | Crystal structure of a polypeptide p53 mutant bound to DNA | | Descriptor: | Cellular tumor antigen p53, ZINC ION, consensus DNA anti-sense strand, ... | | Authors: | Emamzadah, S.T, Tropia, L, Vincenti, I, Falquet, B, Halazonetis, T.D. | | Deposit date: | 2013-09-30 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Reversal of the DNA-Binding-Induced Loop L1 Conformational Switch in an Engineered Human p53 Protein.
J.Mol.Biol., 426, 2014
|
|
8R1F
 
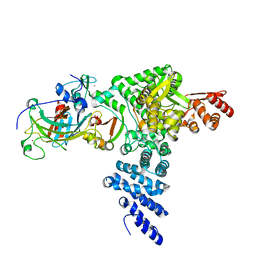 | | Monomeric E6AP-E6-p53 ternary complex | | Descriptor: | Cellular tumor antigen p53, Ubiquitin-like protein SMT3,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Sandate, C.R, Chakraborty, D, Kater, L, Kempf, G, Thoma, N.H. | | Deposit date: | 2023-11-01 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Monomeric E6AP-E6-p53 ternary complex
To Be Published
|
|
8R1G
 
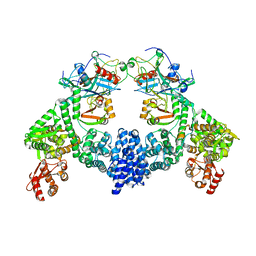 | | Dimeric ternary structure of E6AP-E6-p53 | | Descriptor: | Cellular tumor antigen p53, Ubiquitin-like protein SMT3,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Sandate, C.R, Chakrabory, D, Kater, L, Kempf, G, Thoma, N.H. | | Deposit date: | 2023-11-01 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Monomeric E6AP-E6-p53 ternary complex
To Be Published
|
|
3TS8
 
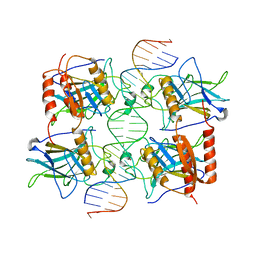 | |
8UQR
 
 | | Crystal structure of the human p53 tetramerization domain | | Descriptor: | Cellular tumor antigen p53 | | Authors: | Wahba, H.M, Sakaguchi, S, Nakagawa, N, Wada, J, Kamada, R, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Highly Similar Tetramerization Domains from the p53 Protein of Different Mammalian Species Possess Varying Biophysical, Functional and Structural Properties.
Int J Mol Sci, 24, 2023
|
|
1A1U
 
 | | SOLUTION STRUCTURE DETERMINATION OF A P53 MUTANT DIMERIZATION DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | P53 | | Authors: | Mccoy, M.A, Stavridi, E.S, Waterman, J.L.F, Wieczorek, A, Opella, S.J, Halezonetis, T.D. | | Deposit date: | 1997-12-16 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Hydrophobic side-chain size is a determinant of the three-dimensional structure of the p53 oligomerization domain.
EMBO J., 16, 1997
|
|
1AIE
 
 | |
1C26
 
 | |
7XZZ
 
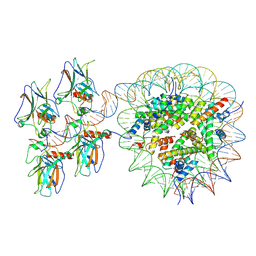 | | Cryo-EM structure of the nucleosome in complex with p53 | | Descriptor: | Cellular tumor antigen p53, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7YFX
 
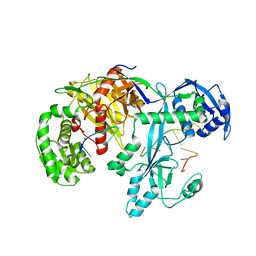 | | Cryo-EM structure of Hili in complex with piRNA | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, piRNA | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
1O9S
 
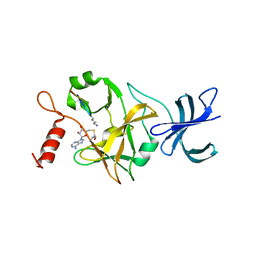 | | Crystal structure of a ternary complex of the human histone methyltransferase SET7/9 | | Descriptor: | GENE FRAGMENT FOR HISTONE H3, HISTONE-LYSINE N-METHYLTRANSFERASE, H3 LYSINE-4 SPECIFIC, ... | | Authors: | Xiao, B, Jing, C, Wilson, J.R, Walker, P.A, Vasisht, N, Kelly, G, Howell, S, Taylor, I.A, Blackburn, G.M, Gamblin, S.J. | | Deposit date: | 2002-12-18 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Catalytic Mechanism of the Human Histone Methyltransferase Set7/9
Nature, 421, 2003
|
|
3Q01
 
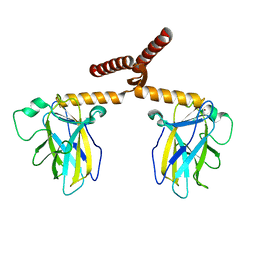 | |
3Q05
 
 | |
3Q06
 
 | |
3SAK
 
 | |
5MR8
 
 | | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K9ac histone peptide | | Descriptor: | E3 ubiquitin-protein ligase TRIM33, Histone H3, ZINC ION | | Authors: | Tallant, C, Savitsky, P, Fedorov, O, Nunez-Alonso, G, Siejka, P, Krojer, T, Williams, E, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K9ac histone peptide
To Be Published
|
|
5NNC
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a diacetylated histone 4 peptide (H3K9ac/K14ac) | | Descriptor: | Bromodomain-containing protein 4, Histone H3 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2017-04-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
