7PCW
 
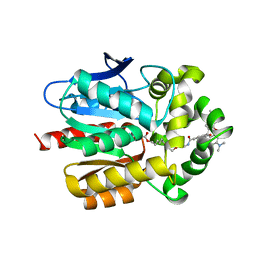 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-M175W LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase, [9-[2-carboxy-5-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]phenyl]-6-(dimethylamino)xanthen-3-ylidene]-dimethyl-azanium | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2021-08-04 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineered HaloTag variants for fluorescence lifetime multiplexing.
Nat.Methods, 19, 2022
|
|
7PCX
 
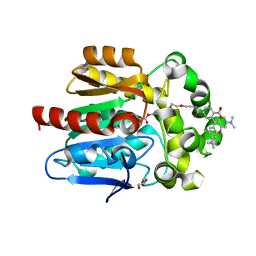 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165W LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2021-08-04 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineered HaloTag variants for fluorescence lifetime multiplexing.
Nat.Methods, 19, 2022
|
|
3Q20
 
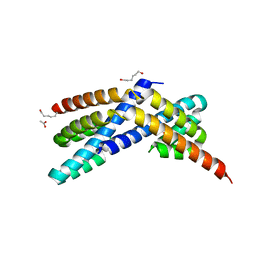 | | Crystal structure of RbcX C103A mutant from Thermosynechococcus elongatus | | Descriptor: | ACETATE ION, HEXANE-1,6-DIOL, RbcX protein | | Authors: | Tarnawski, M, Krzywda, S, Szczepaniak, A, Jaskolski, M. | | Deposit date: | 2010-12-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of the RuBisCO chaperone RbcX from the thermophilic cyanobacterium Thermosynechococcus elongatus
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5DJU
 
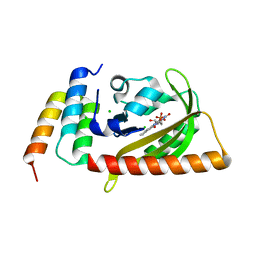 | | Crystal structure of LOV2 (C450A) domain in complex with Zdk3 | | Descriptor: | CHLORIDE ION, Engineered protein, Zdk3 affibody, ... | | Authors: | Tarnawski, M, Wang, H, Yumerefendi, H, Hahn, K.M, Schlichting, I. | | Deposit date: | 2015-09-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | LOVTRAP: an optogenetic system for photoinduced protein dissociation.
Nat.Methods, 13, 2016
|
|
8OVO
 
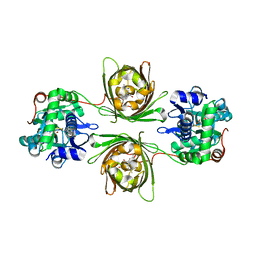 | | X-ray structure of the SF-iGluSnFR-S72A in complex with L-aspartate | | Descriptor: | ASPARTIC ACID, Putative periplasmic binding transport protein,Green fluorescent protein | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the SF-iGluSnFR-S72A in complex with L-aspartate
To Be Published
|
|
8OVP
 
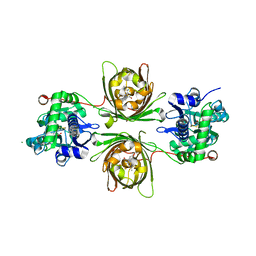 | | X-ray structure of the iAspSnFR in complex with L-aspartate | | Descriptor: | ACETATE ION, ASPARTIC ACID, MAGNESIUM ION, ... | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the SF-iAspSnFR in complex with L-aspartate
To Be Published
|
|
5DJT
 
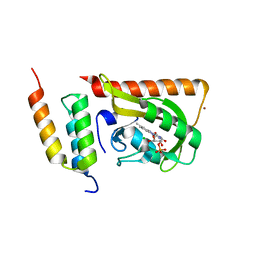 | | Crystal structure of LOV2 (C450A) domain in complex with Zdk2 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Engineered protein, ... | | Authors: | Tarnawski, M, Wang, H, Yumerefendi, H, Hahn, K.M, Schlichting, I. | | Deposit date: | 2015-09-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | LOVTRAP: an optogenetic system for photoinduced protein dissociation.
Nat.Methods, 13, 2016
|
|
5HZH
 
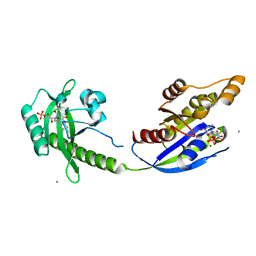 | | Crystal structure of photoinhibitable Rac1 containing C450A mutant LOV2 domain | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tarnawski, M, Dagliyan, O, Chu, P.H, Shirvanyants, D, Dokholyan, N.V, Hahn, K.M, Schlichting, I. | | Deposit date: | 2016-02-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering extrinsic disorder to control protein activity in living cells.
Science, 354, 2016
|
|
5HZI
 
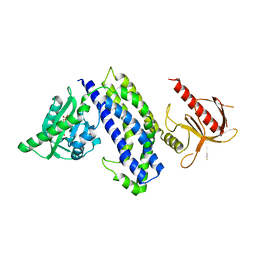 | | Crystal structure of photoinhibitable Intersectin1 containing C450M mutant LOV2 domain | | Descriptor: | FLAVIN MONONUCLEOTIDE, Intersectin-1,NPH1-1,Intersectin-1 | | Authors: | Tarnawski, M, Dagliyan, O, Chu, P.H, Shirvanyants, D, Dokholyan, N.V, Hahn, K.M, Schlichting, I. | | Deposit date: | 2016-02-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering extrinsic disorder to control protein activity in living cells.
Science, 354, 2016
|
|
5HZJ
 
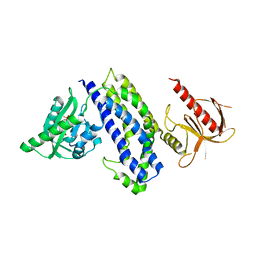 | | Crystal structure of photoinhibitable Intersectin1 containing wildtype LOV2 domain | | Descriptor: | FLAVIN MONONUCLEOTIDE, Intersectin-1,NPH1-1,Intersectin-1 | | Authors: | Tarnawski, M, Dagliyan, O, Chu, P.H, Shirvanyants, D, Dokholyan, N.V, Hahn, K.M, Schlichting, I. | | Deposit date: | 2016-02-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering extrinsic disorder to control protein activity in living cells.
Science, 354, 2016
|
|
5HZK
 
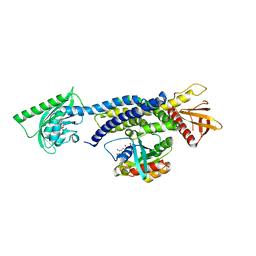 | | Crystal structure of photoinhibitable Intersectin1 containing wildtype LOV2 domain in complex with Cdc42 | | Descriptor: | Cell division control protein 42 homolog, FLAVIN MONONUCLEOTIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tarnawski, M, Dagliyan, O, Chu, P.H, Shirvanyants, D, Dokholyan, N.V, Hahn, K.M, Schlichting, I. | | Deposit date: | 2016-02-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Engineering extrinsic disorder to control protein activity in living cells.
Science, 354, 2016
|
|
8OVN
 
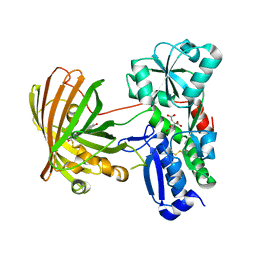 | | X-ray structure of the SF-iGluSnFR-S72A | | Descriptor: | CITRIC ACID, Putative periplasmic binding transport protein,Green fluorescent protein | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the SF-iGluSnFR-S72A
To Be Published
|
|
6Y7B
 
 | | X-ray structure of the Haloalkane dehalogenase HaloTag7 labeled with a chloroalkane-carbopyronine fluorophore substrate | | Descriptor: | 4-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]-2-[3-(dimethylamino)-6-(dimethyl-$l^{4}-azanylidene)-10,10-dimethyl-anthracen-9-yl]benzoic acid, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-02-28 | | Release date: | 2021-03-31 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
6Y7A
 
 | | X-ray structure of the Haloalkane dehalogenase HaloTag7 labeled with a chloroalkane-tetramethylrhodamine fluorophore substrate | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-02-28 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
8B6N
 
 | |
8B6P
 
 | |
8B6S
 
 | | X-ray structure of the haloalkane dehalogenase HaloTag7 fusion to the green fluorescent protein GFP (ChemoG1) labeled with a chloroalkane tetramethylrhodamine fluorophore substrate | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein,Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Hellweg, L, Hiblot, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A general method for the development of multicolor biosensors with large dynamic ranges.
Nat.Chem.Biol., 19, 2023
|
|
6ZCC
 
 | | X-ray structure of the Haloalkane dehalogenase HOB (HaloTag7-based Oligonucleotide Binder) labeled with a chloroalkane-tetramethylrhodamine fluorophore substrate | | Descriptor: | ACETATE ION, CALCIUM ION, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-06-10 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
8B6T
 
 | | X-ray structure of the interface optimized haloalkane dehalogenase HaloTag7 fusion to the green fluorescent protein GFP (ChemoG5-TMR) labeled with a chloroalkane tetramethylrhodamine fluorophore substrate | | Descriptor: | CHLORIDE ION, Green fluorescent protein,Haloalkane dehalogenase, [9-[2-carboxy-5-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]phenyl]-6-(dimethylamino)xanthen-3-ylidene]-dimethyl-azanium | | Authors: | Tarnawski, M, Hellweg, L, Hiblot, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A general method for the development of multicolor biosensors with large dynamic ranges.
Nat.Chem.Biol., 19, 2023
|
|
8B6R
 
 | | X-ray structure of the haloalkane dehalogenase HaloTag7 labeled with a chloroalkane Cyanine3 fluorophore substrate | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Hellweg, L, Hiblot, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A general method for the development of multicolor biosensors with large dynamic ranges.
Nat.Chem.Biol., 19, 2023
|
|
6ZVY
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H-P174R LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineered HaloTag variants for fluorescence lifetime multiplexing.
Nat.Methods, 19, 2022
|
|
7ZIY
 
 | | X-ray structure of the haloalkane dehalogenase HaloTag7 bound to a pentyltrifluoromethanesulfonamide tetramethylrhodamine ligand (TMR-T5) | | Descriptor: | CALCIUM ION, Haloalkane dehalogenase, [9-[2-carboxy-5-[2-[2-[5-(trifluoromethylsulfonylamino)pentoxy]ethoxy]ethylcarbamoyl]phenyl]-6-(dimethylamino)xanthen-3-ylidene]-dimethyl-azanium | | Authors: | Tarnawski, M, Kompa, J, Johnsson, K, Hiblot, J. | | Deposit date: | 2022-04-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exchangeable HaloTag Ligands for Super-Resolution Fluorescence Microscopy.
J.Am.Chem.Soc., 145, 2023
|
|
7ZIZ
 
 | | X-ray structure of the dead variant haloalkane dehalogenase HaloTag7-D106A bound to a pentanol tetramethylrhodamine ligand (TMR-Hy5) | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Kompa, J, Johnsson, K, Hiblot, J. | | Deposit date: | 2022-04-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exchangeable HaloTag Ligands for Super-Resolution Fluorescence Microscopy.
J.Am.Chem.Soc., 145, 2023
|
|
7ZJ0
 
 | | X-ray structure of the haloalkane dehalogenase HaloTag7 bound to a pentylmethanesulfonamide tetramethylrhodamine ligand (TMR-S5) | | Descriptor: | GLYCEROL, Haloalkane dehalogenase, [9-[2-carboxy-5-[2-[2-[5-(methylsulfonylamino)pentoxy]ethoxy]ethylcarbamoyl]phenyl]-6-(dimethylamino)xanthen-3-ylidene]-dimethyl-azanium | | Authors: | Tarnawski, M, Kompa, J, Johnsson, K, Hiblot, J. | | Deposit date: | 2022-04-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exchangeable HaloTag Ligands for Super-Resolution Fluorescence Microscopy.
J.Am.Chem.Soc., 145, 2023
|
|
4HU4
 
 | | Crystal structure of EAL domain of the E. coli DosP - dimeric form | | Descriptor: | Oxygen sensor protein DosP | | Authors: | Tarnawski, M, Barends, T.R.M, Hartmann, E, Schlichting, I. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the catalytic EAL domain of the Escherichia coli direct oxygen sensor.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
