6CVH
 
 | | Identification and biological evaluation of thiazole-based inverse agonists of RORgt | | Descriptor: | Nuclear receptor ROR-gamma, trans-3-({4-(cyclohexylmethyl)-5-[3-(1-methylcyclopropyl)-5-{[(2R)-1,1,1-trifluoropropan-2-yl]carbamoyl}phenyl]-1,3-thiazole-2-carbonyl}amino)cyclobutane-1-carboxylic acid | | Authors: | Spurlino, J, Milligan, C. | | Deposit date: | 2018-03-28 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Identification and biological evaluation of thiazole-based inverse agonists of ROR gamma t.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
3DM6
 
 | | Beta-secretase 1 complexed with statine-based inhibitor | | Descriptor: | 5-[[(2S)-2-[[(3R,4S)-5-(3,5-difluorophenoxy)-3-hydroxy-4-[[3-(methyl-methylsulfonyl-amino)-5-[[(1R)-1-phenylethyl]carbamoyl]phenyl]carbonylamino]pentanoyl]amino]-3-methyl-butanoyl]amino]benzene-1,3-dicarboxylic acid, Beta-secretase 1, ISOPROPYL ALCOHOL | | Authors: | Lindberg, J, Borkakoti, N, Nystrom, S. | | Deposit date: | 2008-06-30 | | Release date: | 2008-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis and SAR of potent statine-based BACE-1 inhibitors: exploration of P1 phenoxy and benzyloxy residues
Bioorg.Med.Chem., 16, 2008
|
|
3UGT
 
 | | Crystal structure of the yeast mitochondrial threonyl-tRNA synthetase - orthorhombic crystal form | | Descriptor: | Threonyl-tRNA synthetase, mitochondrial, ZINC ION | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Cho, C, Soll, D, Simonovic, M. | | Deposit date: | 2011-11-02 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UH0
 
 | | Crystal structure of the yeast mitochondrial threonyl-tRNA synthetase (MST1) in complex with threonyl sulfamoyl adenylate | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, SULFATE ION, Threonyl-tRNA synthetase, ... | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Cho, C, Soll, D, Simonovic, M. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UGQ
 
 | | Crystal structure of the apo form of the yeast mitochondrial threonyl-tRNA synthetase determined at 2.1 Angstrom resolution | | Descriptor: | POTASSIUM ION, SULFATE ION, Threonyl-tRNA synthetase, ... | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Cho, C, Soll, D, Simonovic, M. | | Deposit date: | 2011-11-02 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1T4U
 
 | | Crystal Structure Analysis of a novel Oxyguanidine bound to Thrombin | | Descriptor: | 2-METHANESULFONYL-BENZENESULFONIC ACID 3-METHYL-5-((1-AMIDINOAMINOOXYMETHYL-CYCLOPROPYL)METHYLOXY)-PHENYLESTER, Hirudin IIIA, Prothrombin | | Authors: | Spurlino, J. | | Deposit date: | 2004-04-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxyguanidines. Part 2: Discovery of a novel orally active thrombin inhibitor through structure-based drug design and parallel synthesis
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
1T4V
 
 | | Crystal Structure Analysis of a novel Oxyguanidine bound to Thrombin | | Descriptor: | Hirudin IIIA, N-ALLYL-5-AMIDINOAMINOOXY-PROPYLOXY-3-CHLORO-N-CYCLOPENTYLBENZAMIDE, Prothrombin | | Authors: | Spurlino, J. | | Deposit date: | 2004-04-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxyguanidines. Part 2: Discovery of a novel orally active thrombin inhibitor through structure-based drug design and parallel synthesis
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
5UFO
 
 | | Structure of RORgt bound to | | Descriptor: | (S)-{4-chloro-2-methoxy-3-[4-(methylsulfonyl)phenyl]quinolin-6-yl}(1-methyl-1H-imidazol-5-yl)[6-(trifluoromethyl)pyridin-3-yl]methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J. | | Deposit date: | 2017-01-05 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Identification and structure activity relationships of quinoline tertiary alcohol modulators of ROR gamma t.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5UHI
 
 | | Structure of RORgt bound to | | Descriptor: | (R)-(4-chloro-2-methoxy-3-{[4-(1H-pyrazol-1-yl)phenyl]methyl}quinolin-6-yl)(4-chlorophenyl)(1-methyl-1H-imidazol-5-yl)methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J, Abad, M. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | Identification and structure activity relationships of quinoline tertiary alcohol modulators of ROR gamma t.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5UFR
 
 | | Structure of RORgt bound to | | Descriptor: | (S)-[4-chloro-2-(dimethylamino)-3-phenylquinolin-6-yl](1-methyl-1H-imidazol-5-yl)(pyridin-4-yl)methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J, Abad, M. | | Deposit date: | 2017-01-05 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Identification and structure activity relationships of quinoline tertiary alcohol modulators of ROR gamma t.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
2RDL
 
 | | Hamster Chymase 2 | | Descriptor: | Chymase 2, METHOXYSUCCINYL-ALA-ALA-PRO-ALA-CHLOROMETHYLKETONE INHIBITOR, SULFATE ION | | Authors: | Spurlino, J, Abad, M, Kervinen, J. | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-30 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for elastolytic substrate specificity in rodent alpha-chymases.
J.Biol.Chem., 283, 2008
|
|
1G2K
 
 | | HIV-1 PROTEASE WITH CYCLIC SULFAMIDE INHIBITOR, AHA047 | | Descriptor: | 3-(7-BENZYL-4,5-DIHYDROXY-1,1-DIOXO-3,6-BIS-PHENOXYMETHYL-1L6-[1,2,7]THIADIAZEPAN-2-YLMETHYL)-N-METHYL-BENZAMIDE, PROTEASE RETROPEPSIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2000-10-20 | | Release date: | 2001-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis and comparative molecular field analysis (CoMFA) of symmetric and nonsymmetric cyclic sulfamide HIV-1 protease inhibitors.
J.Med.Chem., 44, 2001
|
|
1G35
 
 | | CRYSTAL STRUCTURE OF HIV-1 PROTEASE IN COMPLEX WITH INHIBITOR, AHA024 | | Descriptor: | 2-[4-(HYDROXY-METHOXY-METHYL)-BENZYL]-7-(4-HYDROXYMETHYL-BENZYL)-1,1-DIOXO-3,6-BIS-PHENOXYMETHYL-1LAMBDA6-[1,2,7]THIADIAZEPANE-4,5-DIOL, HIV-1 PROTEASE | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2000-10-23 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and comparative molecular field analysis (CoMFA) of symmetric and nonsymmetric cyclic sulfamide HIV-1 protease inhibitors.
J.Med.Chem., 44, 2001
|
|
4DPF
 
 | | BACE-1 in complex with a HEA-macrocyclic type inhibitor | | Descriptor: | Beta-secretase 1, N-[(4S,8E,11S)-4-[(1R)-1-hydroxy-2-{[3-(propan-2-yl)benzyl]amino}ethyl]-2,13-dioxo-11-phenyl-6-oxa-3,12-diazabicyclo[12.3.1]octadeca-1(18),8,14,16-tetraen-16-yl]-N-methylmethanesulfonamide | | Authors: | Lindberg, J, Borkakoti, N, Derbyshire, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-07-11 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly potent macrocyclic BACE-1 inhibitors incorporating a hydroxyethylamine core: design, synthesis and X-ray crystal structures of enzyme inhibitor complexes.
Bioorg.Med.Chem., 20, 2012
|
|
4DPI
 
 | | BACE-1 in complex with HEA-macrocyclic inhibitor, MV078512 | | Descriptor: | (4S,8E,11R)-4-[(1R)-1-hydroxy-2-{[3-(propan-2-yl)benzyl]amino}ethyl]-16-methyl-11-phenyl-6-oxa-3,12-diazabicyclo[12.3.1]octadeca-1(18),8,14,16-tetraene-2,13-dione, Beta-secretase 1 | | Authors: | Lindberg, J, Borkakoti, N, Derbyshire, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-07-11 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly potent macrocyclic BACE-1 inhibitors incorporating a hydroxyethylamine core: design, synthesis and X-ray crystal structures of enzyme inhibitor complexes.
Bioorg.Med.Chem., 20, 2012
|
|
4W2E
 
 | | Crystal structure of Elongation Factor 4 (EF4/LepA) bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Lin, J, Steitz, T.A. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-01 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of elongation factor 4 bound to a clockwise ratcheted ribosome.
Science, 345, 2014
|
|
3BZE
 
 | | The human non-classical major histocompatibility complex molecule HLA-E | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | Authors: | Hoare, H.L, Sullivan, L.C, Ely, L.K, Beddoe, T, Henderson, K.N, Lin, J, Clements, C.S, Reid, H.H, Brooks, A.G, Rossjohn, J. | | Deposit date: | 2008-01-17 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Subtle changes in peptide conformation profoundly affect recognition of the non-classical MHC class I molecule HLA-E by the CD94-NKG2 natural killer cell receptors
J.Mol.Biol., 377, 2008
|
|
3CDG
 
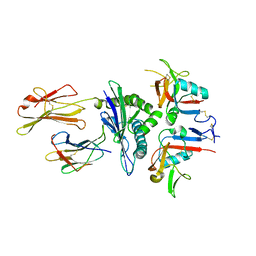 | | Human CD94/NKG2A in complex with HLA-E | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | Authors: | Petrie, E.J, Clements, C.S, Lin, J, Sullivan, L.C, Johnson, D, Huyton, T, Heroux, A, Hoare, H.L, Beddoe, T, Reid, H.H, Wilce, M.C.J, Brooks, A.G, Rossjohn, J. | | Deposit date: | 2008-02-26 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | CD94-NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence
J.Exp.Med., 205, 2008
|
|
4TYB
 
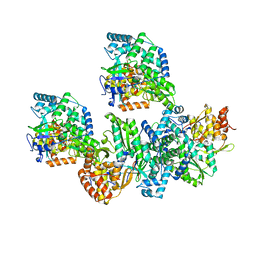 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (2R)-morpholin-4-yl(phenyl)ethanenitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TY9
 
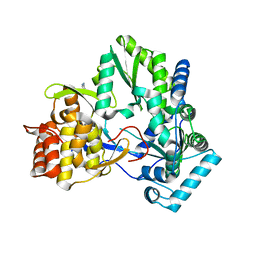 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 5-(trifluoromethyl)pyridin-2-amine, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TYA
 
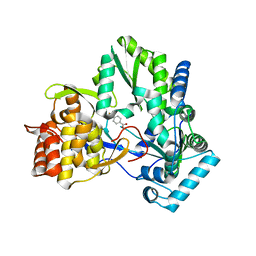 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 4-(trifluoromethyl)benzoic acid, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TXS
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (4-hydroxyphenyl)acetonitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TY8
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 6-methyl-2H-chromen-2-one, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
1VRU
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1VRT
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
