6HPJ
 
 | | Structure of human SRSF1 RRM1 bound to AACAAA RNA | | Descriptor: | Immunoglobulin G-binding protein G,Serine/arginine-rich splicing factor 1, RNA (5'-R(*AP*AP*CP*AP*AP*A)-3') | | Authors: | Allain, F.T.H, Clery, A. | | Deposit date: | 2018-09-21 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of SRSF1 RRM1 bound to RNA reveals an unexpected bimodal mode of interaction and explains its involvement in SMN1 exon7 splicing.
Nat Commun, 12, 2021
|
|
4QU7
 
 | |
4QU6
 
 | |
8BVA
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_114-126 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, L(+)-TARTARIC ACID, POTASSIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-12-02 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_114-126
To Be Published
|
|
8C1J
 
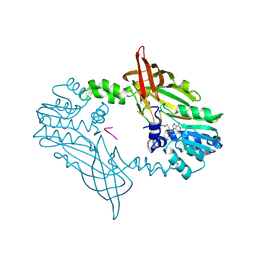 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_18-30 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Protein arginine N-methyltransferase 2, RNA-binding protein Rsf1-like | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_18-30
To Be Published
|
|
1G8Y
 
 | | CRYSTAL STRUCTURE OF THE HEXAMERIC REPLICATIVE HELICASE REPA OF PLASMID RSF1010 | | Descriptor: | REGULATORY PROTEIN REPA | | Authors: | Niedenzu, T, Roeleke, D, Bains, G, Scherzinger, E, Saenger, W. | | Deposit date: | 2000-11-21 | | Release date: | 2001-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010.
J.Mol.Biol., 306, 2001
|
|
3H25
 
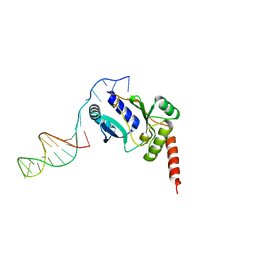 | | Crystal structure of the catalytic domain of primase Repb' in complex with initiator DNA | | Descriptor: | Replication protein B, SINGLE STRANDED INITIATOR DNA (SSIA) | | Authors: | Geibel, S, Banchenko, S, Engel, M, Lanka, E, Saenger, W. | | Deposit date: | 2009-04-14 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of primase RepB' encoded by broad-host-range plasmid RSF1010 that replicates exclusively in leading-strand mode
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3H20
 
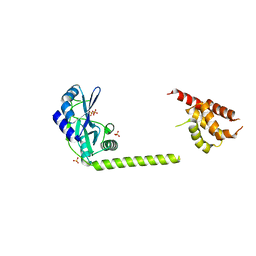 | | Crystal structure of primase RepB' | | Descriptor: | DIPHOSPHATE, Replication protein B, SULFATE ION | | Authors: | Geibel, S, Banchenko, S, Engel, M, Lanka, E, Saenger, W. | | Deposit date: | 2009-04-14 | | Release date: | 2009-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure and function of primase RepB' encoded by broad-host-range plasmid RSF1010 that replicates exclusively in leading-strand mode
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1OLO
 
 | |
2EQP
 
 | | Solution structure of the stn_TNFRSF12A_TNFR domain of Tumor necrosis factor receptor superfamily member 12A precursor | | Descriptor: | Tumor necrosis factor receptor superfamily member 12A | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the stn_TNFRSF12A_TNFR domain of Tumor necrosis factor receptor superfamily member 12A precursor
To be Published
|
|
2M8D
 
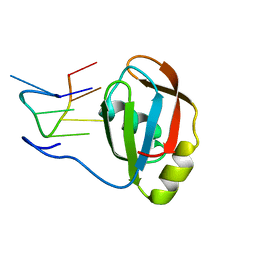 | | Structure of SRSF1 RRM2 in complex with the RNA 5'-UGAAGGAC-3' | | Descriptor: | RNA (5'-R(*UP*GP*AP*AP*GP*GP*AP*C)-3'), Serine/arginine-rich splicing factor 1 | | Authors: | Clery, A, Sinha, R, Anczukow, O, Corrionero, A, Moursy, A, Daubner, G, Valcarcel, J, Krainer, A.R, Allain, F.H.T. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Isolated pseudo-RNA-recognition motifs of SR proteins can regulate splicing using a noncanonical mode of RNA recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2LMI
 
 | |
7ABG
 
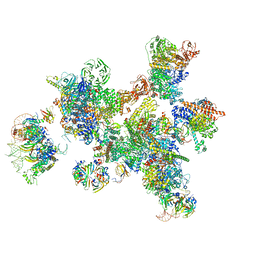 | | Human pre-Bact-1 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Cell division cycle 5-like protein, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
5CIR
 
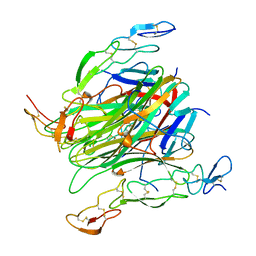 | | Crystal structure of death receptor 4 (DR4; TNFFRSF10A) bound to TRAIL (TNFSF10) | | Descriptor: | CHLORIDE ION, Tumor necrosis factor ligand superfamily member 10, Tumor necrosis factor receptor superfamily member 10A, ... | | Authors: | Sheriff, S. | | Deposit date: | 2015-07-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the death receptor 4-TNF-related apoptosis-inducing ligand (DR4-TRAIL) complex.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
8QO9
 
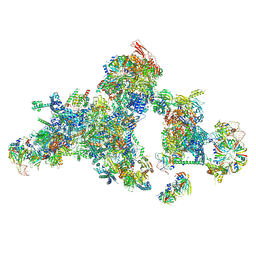 | | Cryo-EM structure of a human spliceosomal B complex protomer | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, MINX pre-mRNA, Microfibrillar-associated protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2023-09-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (5.29 Å) | | Cite: | Cryo-EM analyses of dimerized spliceosomes provide new insights into the functions of B complex proteins.
Embo J., 43, 2024
|
|
4C0O
 
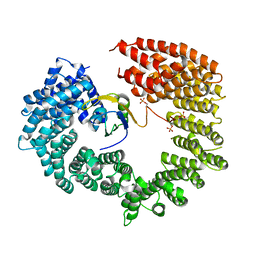 | |
2M7S
 
 | | NMR structure of RNA recognition motif 2 (RRM2) of Homo sapiens splicing factor, arginine/serine-rich 1 | | Descriptor: | Serine/arginine-rich splicing factor 1 | | Authors: | Dutta, S.K, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG), Partnership for T-Cell Biology (TCELL) | | Deposit date: | 2013-04-29 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of RNA recognition motif 2 (RRM2) of Homo sapiens splicing factor, arginine/serine-rich 1
To be Published
|
|
4FHQ
 
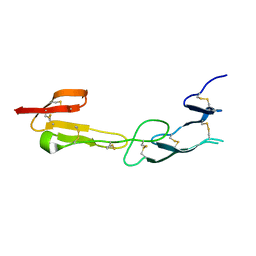 | | Crystal Structure of HVEM | | Descriptor: | Tumor necrosis factor receptor superfamily member 14 | | Authors: | Liu, W, Zhan, C, Patskovsky, Y, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Increased Heterologous Protein Expression in Drosophila S2 Cells for Massive Production of Immune Ligands/Receptors and Structural Analysis of Human HVEM.
Mol Biotechnol, 57, 2015
|
|
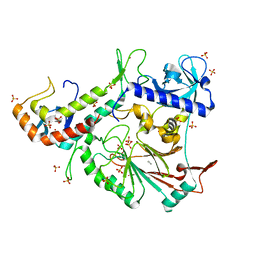
3FLO
 
 | |
7CSQ
 
 | | Solution structure of the complex between p75NTR-DD and TRADD-DD | | Descriptor: | Tumor necrosis factor receptor superfamily member 16, Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Lin, Z, Zhang, N. | | Deposit date: | 2020-08-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of NF-kappa B signaling by the p75 neurotrophin receptor interaction with adaptor protein TRADD through their respective death domains.
J.Biol.Chem., 297, 2021
|
|
7DD1
 
 | |
2KMZ
 
 | | NMR Structure of hFn14 | | Descriptor: | Tumor necrosis factor receptor superfamily member 12A | | Authors: | Pellegrini, M, Willen, L, Perroud, M, Krushinskie, D, Strauch, K, Cuervo, H, Sun, Y, Day, E.S, Schneider, P, Zheng, T.S. | | Deposit date: | 2009-08-10 | | Release date: | 2011-06-29 | | Last modified: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the extracellular domains of human and Xenopus Fn14: implications in the evolution of TWEAK and Fn14 interactions.
Febs J., 280, 2013
|
|
6FAD
 
 | | SR protein kinase 1 (SRPK1) in complex with the RGG-box of HSV1 ICP27 | | Descriptor: | PHOSPHATE ION, SRSF protein kinase 1, mRNA export factor | | Authors: | Tunnicliffe, R.B, Levy, C, Mould, A.P, Mckenzie, E.A, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2017-12-15 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Molecular Mechanism of SR Protein Kinase 1 Inhibition by the Herpes Virus Protein ICP27.
Mbio, 10, 2019
|
|
6BNH
 
 | | Solution NMR structures of BRD4 ET domain with JMJD6 peptide | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, Bromodomain-containing protein 4 | | Authors: | Konuma, T, Yu, D, Zhao, C, Ju, Y, Sharma, R, Ren, C, Zhang, Q, Zhou, M.-M, Zeng, L. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Mechanism of the Oxygenase JMJD6 Recognition by the Extraterminal (ET) Domain of BRD4.
Sci Rep, 7, 2017
|
|
4ZFO
 
 | | J22.9-xi: chimeric mouse/human antibody against human BCMA (CD269) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, J22.9-xi Fab, ... | | Authors: | Marino, S.F, Daumke, O, Olal, D. | | Deposit date: | 2015-04-21 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Potent anti-tumor response by targeting B cell maturation antigen (BCMA) in a mouse model of multiple myeloma.
Mol Oncol, 9, 2015
|
|
