7S5Z
 
 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S61
 
 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S60
 
 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S5Y
 
 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S5X
 
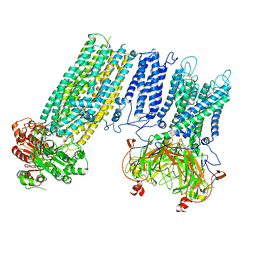 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S5V
 
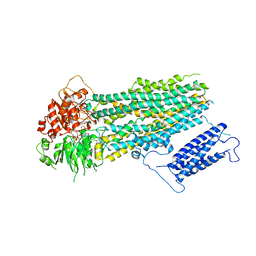 | | Human KATP channel in open conformation, focused on SUR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S5T
 
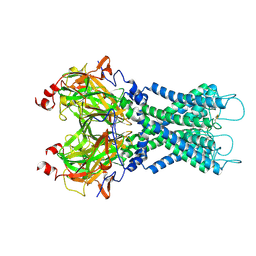 | |
4Y1O
 
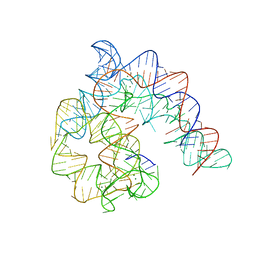 | | Oceanobacillus iheyensis group II intron domain 1 | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, group II intron, ... | | Authors: | Zhao, C, Rajashankar, K.R, Marcia, M, Pyle, A.M. | | Deposit date: | 2015-02-08 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of group II intron domain 1 reveals a template for RNA assembly.
Nat.Chem.Biol., 11, 2015
|
|
4Y1N
 
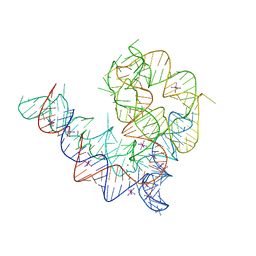 | | Oceanobacillus iheyensis group II intron domain 1 with iridium hexamine | | Descriptor: | IRIDIUM HEXAMMINE ION, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Zhao, C, Rajashankar, K.R, Marcia, M, Pyle, A.M. | | Deposit date: | 2015-02-08 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of group II intron domain 1 reveals a template for RNA assembly.
Nat.Chem.Biol., 11, 2015
|
|
8DDX
 
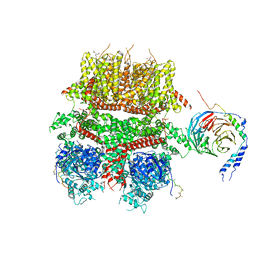 | | cryo-EM structure of TRPM3 ion channel in complex with Gbg in the presence of PIP2, tethered by ALFA-nanobody | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-19 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDW
 
 | |
2A4F
 
 | | Synthesis and Activity of N-Axyl Azacyclic Urea HIV-1 Protease Inhibitors with High Potency Against Multiple Drug Resistant Viral Strains. | | Descriptor: | (5R,6R)-5-BENZYL-6-HYDROXY-2,4-BIS(4-HYDROXY-3-METHOXYBENZYL)-1-[3-(4-HYDROXYPHENYL)PROPANOYL]-1,2,4-TRIAZEPAN-3-ONE, Pol polyprotein | | Authors: | Zhao, C, Sham, H, Sun, M, Lin, S, Stoll, V, Stewart, K.D, Mo, H, Vasavanonda, S, Saldivar, A, McDonald, E. | | Deposit date: | 2005-06-28 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and activity of N-acyl azacyclic urea HIV-1 protease inhibitors with high potency against multiple drug resistant viral strains
Bioorg.Med.Chem.Lett., 15, 2005
|
|
8JZN
 
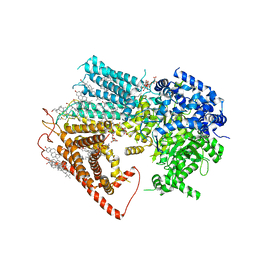 | | Structure of a fungal 1,3-beta-glucan synthase | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, C, You, Z, Chen, D, Hang, J, Wang, Z, Meng, J, Wang, L, Zhao, P, Qiao, J, Yun, C, Bai, L. | | Deposit date: | 2023-07-05 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structure of a fungal 1,3-beta-glucan synthase.
Sci Adv, 9, 2023
|
|
8DDT
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state2 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDR
 
 | | cryo-EM structure of TRPM3 ion channel in the absence of PIP2 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDS
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state1 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDV
 
 | | Cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state4 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-19 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDU
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state3 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-19 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDQ
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of soluble Gbg, focused on channel | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8ED8
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2 and PregS, state 1 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8ED7
 
 | | cryo-EM structure of TRPM3 ion channel in apo state | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, Transient receptor potential cation channel, subfamily M, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-02 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8ED9
 
 | | cryo-EM structure of TRPM3 ion channel in the presence with PIP2 and PregS, state 2 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-09 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
1J0G
 
 | | Solution Structure of Mouse Hypothetical 9.1 kDa Protein, A Ubiquitin-like Fold | | Descriptor: | Hypothetical Protein 1810045K17 | | Authors: | Zhao, C, Kigawa, T, Koshiba, S, Tochio, N, Kobayashi, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mouse Hypothetical 9.1 kDa Protein, A Ubiquitin-like Fold
To be Published
|
|
5HHL
 
 | |
1MXK
 
 | | NMR Structure of HO2-Co(III)bleomycin A(2) Bound to d(GGAAGCTTCC)(2) | | Descriptor: | 5'-D(*GP*GP*AP*AP*GP*CP*TP*TP*CP*C)-3', BLEOMYCIN A2, COBALT (III) ION, ... | | Authors: | Zhao, C, Xia, C, Mao, Q, Forsterling, H, DeRose, E, Antholine, W.E, Subczynski, W.K, Petering, D.H. | | Deposit date: | 2002-10-02 | | Release date: | 2002-10-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structures of HO(2)-Co(III)bleomycin A(2) Bound to d(GAGCTC)(2) and d(GGAAGCTTCC)(2): Structure-Reactivity Relationships of Co and Fe Bleomycins
J.Inorg.Biochem., 91, 2002
|
|
