4GIW
 
 | |
7RD7
 
 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E2P-transition state | | Descriptor: | MAGNESIUM ION, Probable phospholipid-transporting ATPase NEO1, TETRAFLUOROALUMINATE ION | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
7RD6
 
 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E2P state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Probable phospholipid-transporting ATPase NEO1 | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
7RD8
 
 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E1-ATP state | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Probable phospholipid-transporting ATPase NEO1 | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (5.64 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
6WB9
 
 | | Structure of the S. cerevisiae ER membrane complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the ER membrane complex, a transmembrane-domain insertase.
Nature, 584, 2020
|
|
5UN0
 
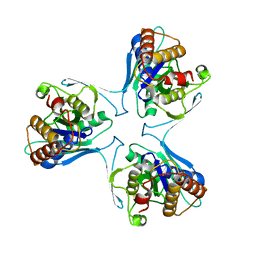 | | Crystal Structure of Mycobacterium Tuberculosis Proteasome-assembly chaperone homologue Rv2125 | | Descriptor: | proteasome assembly chaperone 2 (PAC2) homologue Rv2125 | | Authors: | Bai, L, Jastrab, J.B, Hu, K, Yu, H, Darwin, K.H, Li, H. | | Deposit date: | 2017-01-30 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Mycobacterium tuberculosis Homologues of the Eukaryotic Proteasome Assembly Chaperone 2 (PAC2).
J. Bacteriol., 199, 2017
|
|
5IET
 
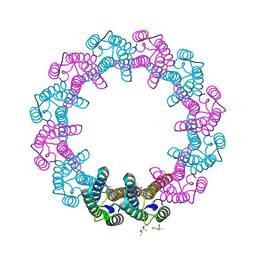 | | Crystal Structure of Mycobacterium Tuberculosis ATP-independent Proteasome activator | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Bacterial proteasome activator, SULFATE ION | | Authors: | Bai, L, Hu, K, Wang, T, Jastrab, J.B, Darwin, K.H, Li, H. | | Deposit date: | 2016-02-25 | | Release date: | 2016-03-30 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural analysis of the dodecameric proteasome activator PafE in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IEU
 
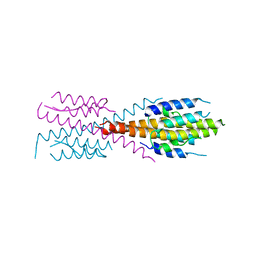 | | Crystal Structure of Mycobacterium Tuberculosis ATP-independent Proteasome Activator Tetramer | | Descriptor: | Bacterial proteasome activator | | Authors: | Bai, L, Hu, K, Wang, T, Jastrab, J.B, Darwin, K.H, Li, H. | | Deposit date: | 2016-02-25 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of the dodecameric proteasome activator PafE in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8K3U
 
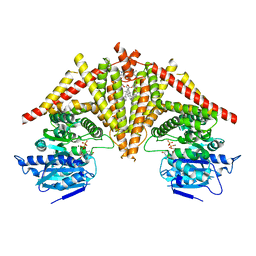 | | S. cerevisiae Chs1 in complex with UDP and GlcNAc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin synthase 1, ... | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase
Nat Commun, 14, 2023
|
|
8K3R
 
 | | S. cerevisiae Chs1 in apo state incubated with GlcNAc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1 | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase
Nat Commun, 14, 2023
|
|
8K3P
 
 | | S. cerevisiae Chs1 in complex with polyoxin B | | Descriptor: | (2S)-2-[[(2S,3S,4S)-5-aminocarbonyloxy-2-azanyl-3,4-bis(oxidanyl)pentanoyl]amino]-2-[(2R,3S,4R,5R)-5-[5-(hydroxymethyl)-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]ethanoic acid, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1 | | Authors: | Bai, L, Chen, D.D. | | Deposit date: | 2023-07-16 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase
Nat Commun, 14, 2023
|
|
8K3X
 
 | | S. cerevisiae Chs1 in complex with Nikkomycin Z | | Descriptor: | (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), Chitin synthase 1 | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase
Nat Commun, 14, 2023
|
|
8K3Q
 
 | | S. cerevisiae Chs1 in apo state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1 | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-16 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase
Nat Commun, 14, 2023
|
|
8K3W
 
 | | S. cerevisiae Chs1 in complex with UDP-GlcNAc and GlcNAc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1, MAGNESIUM ION, ... | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase
Nat Commun, 14, 2023
|
|
8K3V
 
 | | S. cerevisiae Chs1 in complex with UDP-GlcNAc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1, MAGNESIUM ION, ... | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase
Nat Commun, 14, 2023
|
|
8K3T
 
 | | S. cerevisiae Chs1 in complex with UDP | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1, MAGNESIUM ION, ... | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase
Nat Commun, 14, 2023
|
|
6P2R
 
 | | Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor | | Descriptor: | (3R)-3,31-dimethyl-7,11,15,19,23,27-hexamethylidenedotriacont-31-en-1-yl dihydrogen phosphate, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the eukaryotic protein O-mannosyltransferase Pmt1-Pmt2 complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6P28
 
 | |
6P25
 
 | | Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor and a peptide acceptor | | Descriptor: | (3R)-3,31-dimethyl-7,11,15,19,23,27-hexamethylidenedotriacont-31-en-1-yl dihydrogen phosphate, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the eukaryotic protein O-mannosyltransferase Pmt1-Pmt2 complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6PSY
 
 | | Cryo-EM structure of S. cerevisiae Drs2p-Cdc50p in the autoinhibited apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell division control protein 50, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2019-07-14 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p.
Nat Commun, 10, 2019
|
|
6PSX
 
 | | Cryo-EM structure of S. cerevisiae Drs2p-Cdc50p in the PI4P-activated form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell division control protein 50, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2019-07-14 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p.
Nat Commun, 10, 2019
|
|
7VO4
 
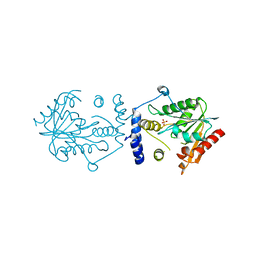 | |
7VO5
 
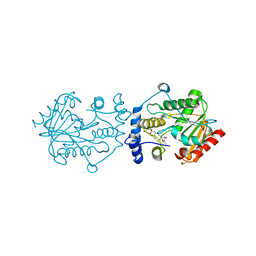 | | Pimaricin type I PKS thioesterase domain (holo Pim TE) | | Descriptor: | (1R,3S,5E,7S,11R,13E,15E,17E,19E,21R,23S,24R,25S)-11,24-dimethyl-1,3,7,21,25-pentakis(oxidanyl)-10,27-dioxabicyclo[21.3.1]heptacosa-5,13,15,17,19-pentaen-9-one, ScnS4 | | Authors: | Bai, L, Zhou, Y. | | Deposit date: | 2021-10-12 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mechanistic Insights into Chain Release of the Polyene PKS Thioesterase Domain
Acs Catalysis, 12, 2022
|
|
6C26
 
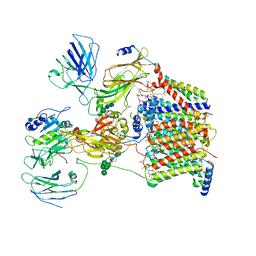 | | The Cryo-EM structure of a eukaryotic oligosaccharyl transferase complex | | Descriptor: | (4R,7R)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(undecanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphadocosan-1-aminium, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2018-01-06 | | Release date: | 2018-01-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The atomic structure of a eukaryotic oligosaccharyltransferase complex.
Nature, 555, 2018
|
|
7KY7
 
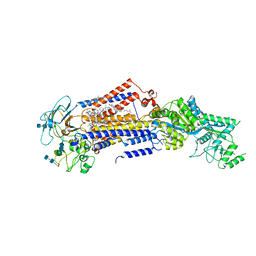 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the apo E1 state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, CHOLESTEROL, ... | | Authors: | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-06 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
