4X3K
 
 | |
4X3U
 
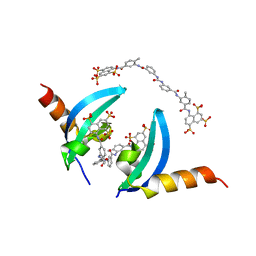 | | Crystal structure of chromobox homolog 7 (CBX7) chromodomain with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Chromobox protein homolog 7 | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain.
Chem.Biol., 22, 2015
|
|
4X3T
 
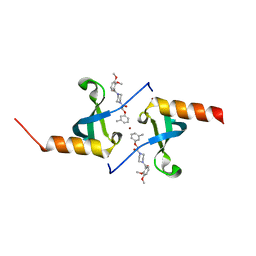 | | Crystal structure of chromobox homolog 7 (CBX7) chromodomain with MS37452 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(2,3-dimethoxybenzoyl)piperazin-1-yl]-2-(3-methylphenoxy)ethanone, Chromobox protein homolog 7, ... | | Authors: | Ren, C, Jakoncic, J, Zhou, M.M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain.
Chem.Biol., 22, 2015
|
|
4X3S
 
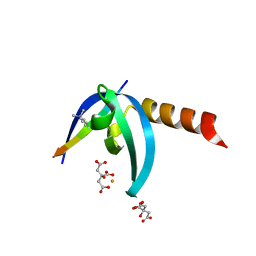 | | Crystal structure of chromobox homology 7 (CBX7) with SETDB1-1170me3 Peptide | | Descriptor: | CITRIC ACID, Chromobox protein homolog 7, FE (III) ION, ... | | Authors: | Ren, C, Plotnikov, A.N, Zhou, M.M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain.
Chem.Biol., 22, 2015
|
|
5AUS
 
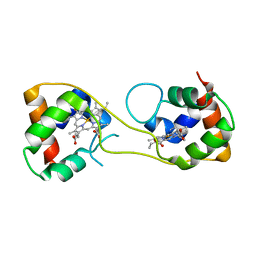 | | Hydrogenobacter thermophilus cytochrome c552 dimer formed by domain swapping at C-terminal region | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Ren, C, Nagao, S, Yamanaka, M, Komori, H, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oligomerization enhancement and two domain swapping mode detection for thermostable cytochrome c552via the elongation of the major hinge loop.
Mol Biosyst, 11, 2015
|
|
5AUR
 
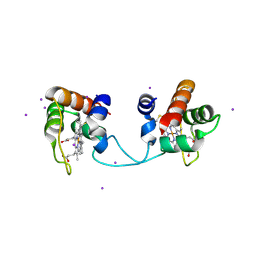 | | Hydrogenobacter thermophilus cytochrome c552 dimer formed by domain swapping at N-terminal region | | Descriptor: | Cytochrome c-552, HEME C, IODIDE ION | | Authors: | Ren, C, Nagao, S, Yamanaka, M, Kamikubo, H, Komori, H, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Oligomerization enhancement and two domain swapping mode detection for thermostable cytochrome c552via the elongation of the major hinge loop.
Mol Biosyst, 11, 2015
|
|
6DNE
 
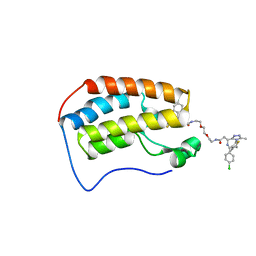 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS660 | | Descriptor: | Bromodomain-containing protein 4, N,N'-[ethane-1,2-diylbis(oxyethane-2,1-diyl)]bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.958 Å) | | Cite: | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DJC
 
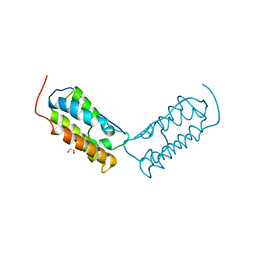 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS645 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,N'-(decane-1,10-diyl)bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2018-05-25 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5EJW
 
 | | Crystal structure of chromobox homolog 7 (CBX7) chromodomain with MS351 | | Descriptor: | (1~{R})-2-[2-azanylidene-3-[(2-methylphenyl)methyl]benzimidazol-1-yl]-1-(3,4-dichlorophenyl)ethanol, Chromobox protein homolog 7 | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Discovery of Selective Antagonists for the Chromodomain of Polycomb Repressive Protein CBX7.
Acs Med.Chem.Lett., 7, 2016
|
|
6BNH
 
 | | Solution NMR structures of BRD4 ET domain with JMJD6 peptide | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, Bromodomain-containing protein 4 | | Authors: | Konuma, T, Yu, D, Zhao, C, Ju, Y, Sharma, R, Ren, C, Zhang, Q, Zhou, M.-M, Zeng, L. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Mechanism of the Oxygenase JMJD6 Recognition by the Extraterminal (ET) Domain of BRD4.
Sci Rep, 7, 2017
|
|
