3ZNI
 
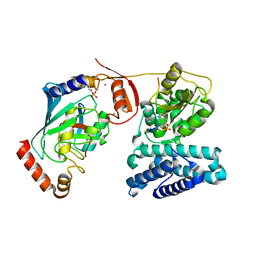 | | Structure of phosphoTyr363-Cbl-b - UbcH5B-Ub - ZAP-70 peptide complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE CBL-B, ... | | Authors: | Dou, H, Buetow, L, Sibbet, G.J, Cameron, K, Huang, D.T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Essentiality of a Non-Ring Element in Priming Donor Ubiquitin for Catalysis by a Monomeric E3.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2VKI
 
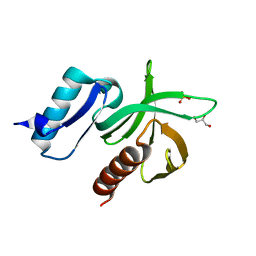 | | Structure of the PDK1 PH domain K465E mutant | | Descriptor: | 3-PHOPSHOINOSITIDE DEPENDENT PROTEIN KINASE 1, GLYCEROL, SULFATE ION | | Authors: | Komander, D, Bayascas, J.R, Deak, M, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2007-12-19 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutation of the Pdk1 Ph Domain Inhibits Protein Kinase B/Akt, Leading to Small Size and Insulin Resistance.
Mol.Cell.Biol., 28, 2008
|
|
4FKD
 
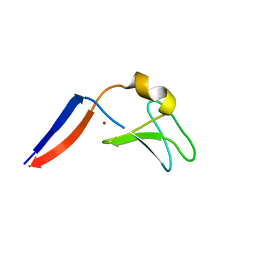 | | Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta | | Descriptor: | Protein kinase C theta type, ZINC ION | | Authors: | Rahman, G.M, Shanker, S, Lewin, N.E, Prasad, B.V.V, Blumberg, P.M, Das, J. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.633 Å) | | Cite: | Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta.
Biochem.J., 451, 2013
|
|
8GCY
 
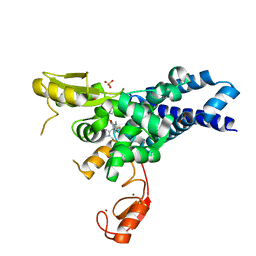 | | Co-crystal structure of CBL-B in complex with N-Aryl isoindolin-1-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Kimani, S, Zeng, H, Dong, A, Li, Y, Santhakumar, V, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition.
Commun Biol, 6, 2023
|
|
3PFV
 
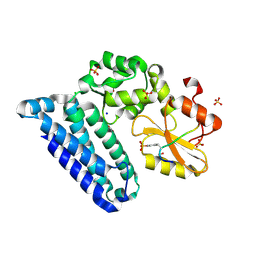 | | Crystal structure of Cbl-b TKB domain in complex with EGFR pY1069 peptide | | Descriptor: | 1,2-ETHANEDIOL, 11-meric peptide from Epidermal growth factor receptor, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Guo, K, Cooper, C.D.O, Ayinampudi, V, Krojer, T, Muniz, J.R.C, Vollmar, M, Canning, P, Gileadi, O, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of Cbl-b TKB domain in complex with EGFR pY1069 peptide
To be Published
|
|
3VGO
 
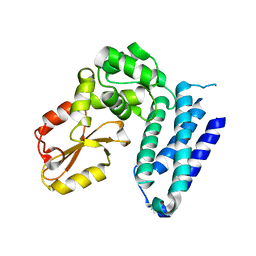 | |
6MHF
 
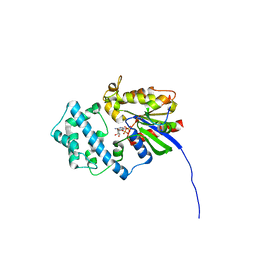 | | Galphai3 co-crystallized with GIV/Girdin | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Girdin, ... | | Authors: | Rees, S.D, Kalogriopoulos, N.A, Ngo, T, Kopcho, N, Ilatovskiy, A, Sun, N, Komives, E, Chang, G, Ghosh, P, Kufareva, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for GPCR-independent activation of heterotrimeric Gi proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4Q9Z
 
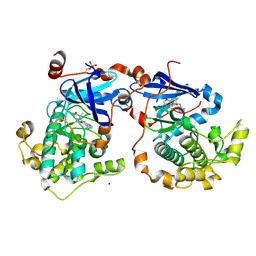 | | Human Protein Kinase C Theta in Complex with Compound35 ((1R)-9-(AZETIDIN-3-YLAMINO)-1,8-DIMETHYL-3,5-DIHYDRO[1,2,4]TRIAZINO[3,4-C][1,4]BENZOXAZIN-2(1H)-ONE) | | Descriptor: | (1R)-9-(azetidin-3-ylamino)-1,8-dimethyl-3,5-dihydro[1,2,4]triazino[3,4-c][1,4]benzoxazin-2(1H)-one, HUMAN PROTEIN KINASE C THETA, SODIUM ION | | Authors: | Argiriadi, M.A, George, D.M. | | Deposit date: | 2014-05-02 | | Release date: | 2014-07-02 | | Last modified: | 2015-01-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Selective and Orally Bioavailable Protein Kinase C theta (PKC theta ) Inhibitors from a Fragment Hit.
J.Med.Chem., 58, 2015
|
|
4RA5
 
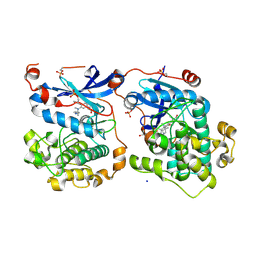 | | Human Protein Kinase C THETA IN COMPLEX WITH LIGAND COMPOUND 11a (6-[(1,3-Dimethyl-azetidin-3-yl)-methyl-amino]-4(R)-methyl-7-phenyl-2,10-dihydro-9-oxa-1,2,4a-triaza-phenanthren-3-one) | | Descriptor: | (1R)-9-[(1,3-dimethylazetidin-3-yl)(methyl)amino]-1-methyl-8-phenyl-3,5-dihydro[1,2,4]triazino[3,4-c][1,4]benzoxazin-2(1H)-one, 1,2-ETHANEDIOL, HUMAN PROTEIN KINASE C THETA, ... | | Authors: | Argiriadi, M.A, George, D.M. | | Deposit date: | 2014-09-09 | | Release date: | 2014-10-08 | | Last modified: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Optimized Protein Kinase C theta (PKC theta ) Inhibitors Reveal Only Modest Anti-inflammatory Efficacy in a Rodent Model of Arthritis.
J.Med.Chem., 58, 2015
|
|
1W1G
 
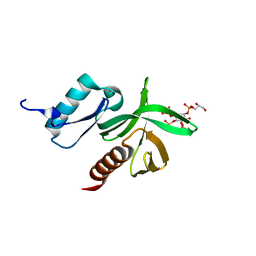 | | Crystal Structure of the PDK1 Pleckstrin Homology (PH) domain bound to DiC4-phosphatidylinositol (3,4,5)-trisphosphate | | Descriptor: | (2R)-3-{[(S)-{[(2S,3R,5S,6S)-2,6-DIHYDROXY-3,4,5-TRIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-(1-HYDROXYBUTOXY)PROPYL BUTYRATE, 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1 | | Authors: | Komander, D, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2004-06-21 | | Release date: | 2004-11-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Insights Into the Regulation of Pdk1 by Phosphoinositides and Inositol Phosphates
Embo J., 23, 2004
|
|
1W1H
 
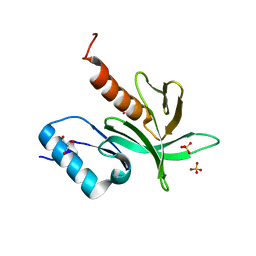 | | Crystal Structure of the PDK1 Pleckstrin Homology (PH) domain | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, GLYCEROL, SULFATE ION | | Authors: | Komander, D, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2004-06-21 | | Release date: | 2004-11-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Insights Into the Regulation of Pdk1 by Phosphoinositides and Inositol Phosphates
Embo J., 23, 2004
|
|
1W1D
 
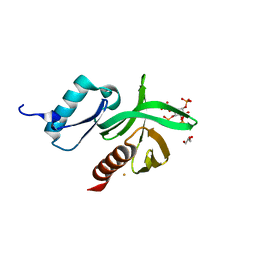 | | Crystal Structure of the PDK1 Pleckstrin Homology (PH) domain bound to Inositol (1,3,4,5)-tetrakisphosphate | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, GLYCEROL, GOLD ION, ... | | Authors: | Komander, D, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2004-06-21 | | Release date: | 2004-11-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights Into the Regulation of Pdk1 by Phosphoinositides and Inositol Phosphates
Embo J., 23, 2004
|
|
5AXI
 
 | | Crystal structure of Cbl-b TKB domain in complex with Cblin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cblin, ... | | Authors: | Ohno, A, Maita, N, Ochi, A, Nakao, R, Nikawa, T. | | Deposit date: | 2015-07-29 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the TKB domain of ubiquitin ligase Cbl-b complexed with its small inhibitory peptide, Cblin
Arch.Biochem.Biophys., 594, 2016
|
|
5D9H
 
 | | Crystal structure of SPAK (STK39) dimer in the basal activity state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, STE20/SPS1-related proline-alanine-rich protein kinase, ... | | Authors: | Taylor, C.A, Juang, Y.C, Goldsmith, E.J, Cobb, M.H. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Domain-Swapping Switch Point in Ste20 Protein Kinase SPAK.
Biochemistry, 54, 2015
|
|
5DBX
 
 | | Crystal structure of murine SPAK(T243D) in complex with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, STE20/SPS1-related proline-alanine-rich protein kinase | | Authors: | Juang, Y.-C. | | Deposit date: | 2015-08-22 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Domain-Swapping Switch Point in Ste20 Protein Kinase SPAK.
Biochemistry, 54, 2015
|
|
5F9E
 
 | | Structure of Protein Kinase C theta with compound 10: 2,2-dimethyl-7-(2-oxidanylidene-3~{H}-imidazo[4,5-b]pyridin-1-yl)-1-(phenylmethyl)-3~{H}-quinazolin-4-one | | Descriptor: | 2,2-dimethyl-7-(2-oxidanylidene-3~{H}-imidazo[4,5-b]pyridin-1-yl)-1-(phenylmethyl)-3~{H}-quinazolin-4-one, Protein kinase C theta type | | Authors: | Klein, M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-05-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of 1,7-disubstituted-2,2-dimethyl-2,3-dihydroquinazolin-4(1H)-ones as potent and selective PKC theta inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
8QNH
 
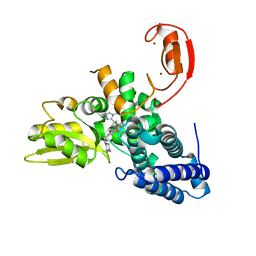 | | Crystal structure of the E3 ubiquitin ligase Cbl-b with an allosteric inhibitor (WO2020264398 Ex23) | | Descriptor: | 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Discovery of a Novel Benzodiazepine Series of Cbl-b Inhibitors for the Enhancement of Antitumor Immunity.
Acs Med.Chem.Lett., 14, 2023
|
|
8QNG
 
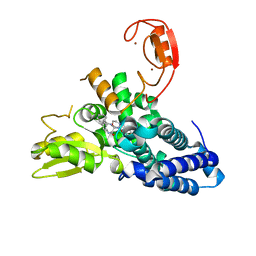 | |
8QNI
 
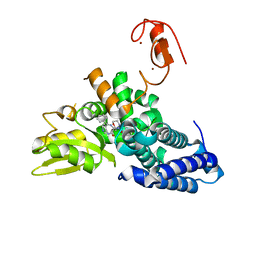 | |
8QTJ
 
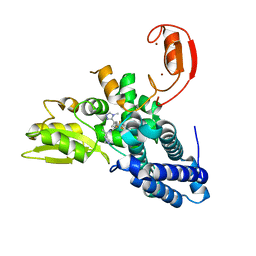 | | Crystal structure of Cbl-b in complex with an allosteric inhibitor (compound 30) | | Descriptor: | 3-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-1-[(1~{R})-1-(1-methylpyrazol-4-yl)ethyl]-5-(trifluoromethyl)pyridin-2-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QTH
 
 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 8) | | Descriptor: | 1-methyl-5-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-3-(trifluoromethyl)-7H-pyrrolo[2,3-b]pyridin-6-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QTG
 
 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 9) | | Descriptor: | 3-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-5-(trifluoromethyl)-1~{H}-pyridin-2-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QTK
 
 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 31) | | Descriptor: | 3-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-1-[(1S)-1-(1-methylpyrazol-4-yl)ethyl]-5-(trifluoromethyl)pyridin-2-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.873 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
