4TX6
 
 | | AfChiA1 in complex with compound 1 | | Descriptor: | 3-(2-methoxyphenyl)-6-methyl[1,2]oxazolo[5,4-d]pyrimidin-4(5H)-one, Class III chitinase ChiA1, PHOSPHATE ION | | Authors: | van Aalten, D.M.F. | | Deposit date: | 2014-07-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Screening-based discovery of Aspergillus fumigatus plant-type chitinase inhibitors
FEBS Lett, 588, 2014
|
|
4EZA
 
 | | Crystal structure of the atypical phosphoinositide (aPI) binding domain of IQGAP2 | | Descriptor: | Ras GTPase-activating-like protein IQGAP2 | | Authors: | Van Aalten, D.M.F, Dixon, M.J, Gray, A, Schenning, M, Agacan, M, Leslie, N.R, Downes, C.P, Batty, I.H, Nedyalkova, L, Tempel, W, Tong, Y, Zhong, N, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-02 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | IQGAP Proteins Reveal an Atypical Phosphoinositide (aPI) Binding Domain with a Pseudo C2 Domain Fold.
J.Biol.Chem., 287, 2012
|
|
1E2X
 
 | | FadR, fatty acid responsive transcription factor from E. coli | | Descriptor: | FATTY ACID METABOLISM REGULATOR PROTEIN, SULFATE ION | | Authors: | Van Aalten, D.M.F, Dirusso, C.C, Knudsen, J, Wierenga, R.K. | | Deposit date: | 2000-05-30 | | Release date: | 2000-12-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Fadr, a Fatty Acid-Responsive Transcription Factor with a Novel Acyl Coenzyme A-Binding Fold
Embo J., 19, 2000
|
|
1H9G
 
 | | FadR, FATTY ACID RESPONSIVE TRANSCRIPTION FACTOR FROM E. COLI, in complex with myristoyl-CoA | | Descriptor: | COENZYME A, FATTY ACID METABOLISM REGULATOR PROTEIN, MYRISTIC ACID | | Authors: | Van Aalten, D.M.F, Dirusso, C.C, Knudsen, J. | | Deposit date: | 2001-03-09 | | Release date: | 2001-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structural Basis of Acyl Coenzyme A-Dependent Regulation of the Transcription Factor Fadr
Embo J., 20, 2001
|
|
1H9T
 
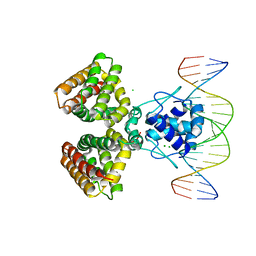 | | FADR, FATTY ACID RESPONSIVE TRANSCRIPTION FACTOR FROM E. COLI IN COMPLEX WITH FADB OPERATOR | | Descriptor: | 5'-D(*CP*AP*TP*CP*TP*GP*GP*TP*AP*CP*GP*AP* CP*CP*AP*GP*AP*TP*C)-3', 5'-D(*GP*AP*TP*CP*TP*GP*GP*TP*CP*GP*TP*AP* CP*CP*AP*GP*AP*TP*G)-3', CHLORIDE ION, ... | | Authors: | Van Aalten, D.M.F, Dirusso, C.C, Knudsen, J. | | Deposit date: | 2001-03-19 | | Release date: | 2001-04-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The Structural Basis of Acyl Coenzyme A-Dependent Regulation of the Transcription Factor Fadr
Embo J., 20, 2001
|
|
1E15
 
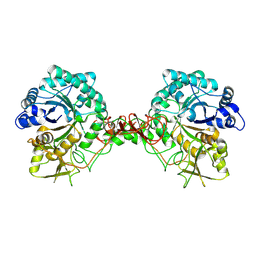 | | Chitinase B from Serratia Marcescens | | Descriptor: | CHITINASE B | | Authors: | Van Aalten, D.M.F, Synstad, B, Brurberg, M.B, Hough, E, Riise, B.W, Eijsink, V.G.H, Wierenga, R.K. | | Deposit date: | 2000-04-18 | | Release date: | 2000-08-18 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Two-Domain Chitotriosidase from Serratia Marcescens at 1.9 Angstrom Resoltuion
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1CW9
 
 | | DNA DECAMER WITH AN ENGINEERED CROSSLINK IN THE MINOR GROOVE | | Descriptor: | 5'-D(*CP*CP*AP*GP*(G47)P*CP*CP*TP*GP*G)-3', CALCIUM ION | | Authors: | van Aalten, D.M.F, Erlanson, D.A, Verdine, G.L, Joshua-Tor, L. | | Deposit date: | 1999-08-26 | | Release date: | 1999-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A structural snapshot of base-pair opening in DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1D7E
 
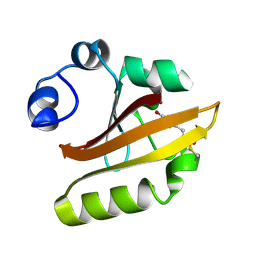 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 1999-10-17 | | Release date: | 2000-03-31 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Conformational substates in different crystal forms of the photoactive yellow protein--correlation with theoretical and experimental flexibility.
Protein Sci., 9, 2000
|
|
1GSV
 
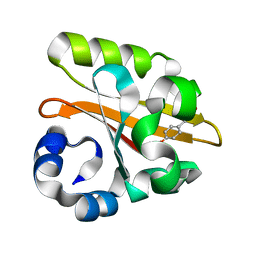 | | Crystal structure of the P65 crystal form of photoactive yellow protein G47S mutant | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-08 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1GSX
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN G47S/G51S MUTANT | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1GSW
 
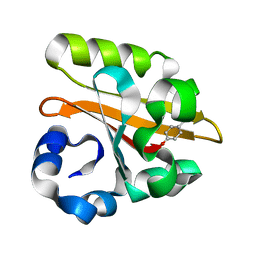 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN G51S MUTANT | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1HBK
 
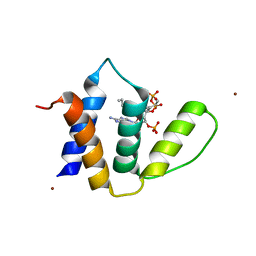 | | Acyl-CoA binding protein from Plasmodium falciparum | | Descriptor: | ACYL-COA BINDING PROTEIN, COENZYME A, MYRISTIC ACID, ... | | Authors: | van Aalten, D.M.F. | | Deposit date: | 2001-04-16 | | Release date: | 2001-05-15 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding site differences revealed by crystal structures of Plasmodium falciparum and bovine acyl-CoA binding protein.
J. Mol. Biol., 309, 2001
|
|
1KOU
 
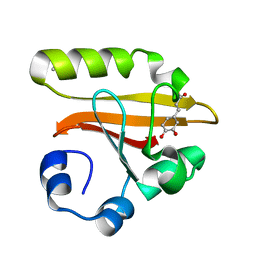 | | Crystal Structure of the Photoactive Yellow Protein Reconstituted with Caffeic Acid at 1.16 A Resolution | | Descriptor: | CAFFEIC ACID, N-BUTANE, PHOTOACTIVE YELLOW PROTEIN | | Authors: | van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2001-12-22 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the photoactive yellow protein reconstituted with caffeic acid at 1.16 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5ZVP
 
 | | Aspergillus fumigatus Rho1 F25N | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rho GTPase Rho1, ... | | Authors: | Wei, W.F, Van Aalten, D.M.F. | | Deposit date: | 2018-05-12 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Aspergillus fumigateurs Rho1 F25N
To Be Published
|
|
7P5O
 
 | |
6Z2F
 
 | |
3ZJ0
 
 | | The human O-GlcNAcase C-terminal domain is a pseudo histone acetyltransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, ACETYLTRANSFERASE, ... | | Authors: | Rao, F.V, Schuettelkopf, A.W, Dorfmueller, H.C, Ferenbach, A.T, Navratilova, I, van Aalten, D.M.F. | | Deposit date: | 2013-01-15 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Bacterial Putative Acetyltransferase Defines the Fold of the Human O-Glcnacase C-Terminal Domain.
Open Biol., 3, 2013
|
|
4WPG
 
 | | Group A Streptococcus GacA is an essential dTDP-4-dehydrorhamnose reductase (RmlD) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, ... | | Authors: | van der Beek, S.L, Le Breton, Y, Ferenbach, A.T, van Aalten, D.M.F, Navratilova, I, McIver, K, van Sorge, N.M, Dorfmueller, H.C. | | Deposit date: | 2014-10-18 | | Release date: | 2015-09-09 | | Last modified: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | GacA is essential for Group A Streptococcus and defines a new class of monomeric dTDP-4-dehydrorhamnose reductases (RmlD).
Mol.Microbiol., 98, 2015
|
|
4TXE
 
 | | ScCTS1 in complex with compound 5 | | Descriptor: | (2S)-1-(2,3-dihydro-1H-inden-2-ylamino)-3-(3,4-dimethylphenoxy)propan-2-ol, Endochitinase | | Authors: | Schuettelkopf, A.W, van Aalten, D.M.F. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Screening-based discovery of Aspergillus fumigatus plant-type chitinase inhibitors
FEBS Lett, 588, 2014
|
|
3GNI
 
 | | Structure of STRAD and MO25 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, Protein Mo25, ... | | Authors: | Zeqiraj, E, Goldie, S, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2009-03-17 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | ATP and MO25alpha regulate the conformational state of the STRADalpha pseudokinase and activation of the LKB1 tumour suppressor
Plos Biol., 7, 2009
|
|
6EOU
 
 | |
7OYL
 
 | | Phosphoglucose isomerase of Aspergillus fumigatus in complexed with Glucose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raimi, O.G, Yan, K, Fang, W, van Aalten, D.M.F. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Phosphoglucose Isomerase Is Important for Aspergillus fumigatus Cell Wall Biogenesis.
Mbio, 13, 2022
|
|
7PJC
 
 | |
7PIZ
 
 | |
7PPR
 
 | |
