7M2K
 
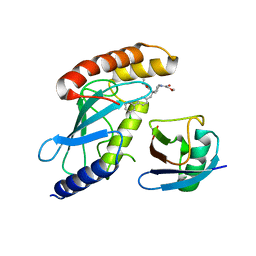 | | CDC34A-Ubiquitin-2ab inhibitor complex | | Descriptor: | 4-[(3',5'-dichloro[1,1'-biphenyl]-4-yl)methyl]-N-ethyl-1-(methoxyacetyl)piperidine-4-carboxamide, Ubiquitin, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, St-Cyr, D, Tyers, M, Sicheri, F. | | Deposit date: | 2021-03-16 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Identification and optimization of molecular glue compounds that inhibit a noncovalent E2 enzyme-ubiquitin complex.
Sci Adv, 7, 2021
|
|
1NEX
 
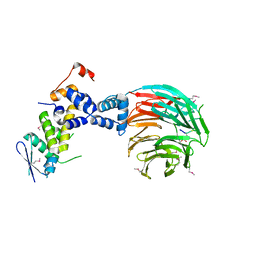 | | Crystal Structure of ScSkp1-ScCdc4-CPD peptide complex | | Descriptor: | CDC4 protein, Centromere DNA-binding protein complex CBF3 subunit D, GLL(TPO)PPQSG | | Authors: | Orlicky, S, Tang, X, Willems, A, Tyers, M, Sicheri, F. | | Deposit date: | 2002-12-12 | | Release date: | 2003-02-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Phosphodependent Substrate Selection and
Orientation by the SCFCdc4 Ubiquitin Ligase
Cell(Cambridge,Mass.), 112, 2003
|
|
4MDK
 
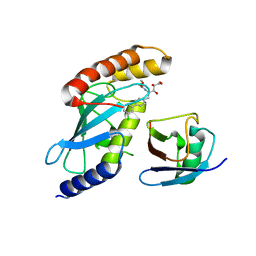 | | Cdc34-ubiquitin-CC0651 complex | | Descriptor: | 4,5-dideoxy-5-(3',5'-dichlorobiphenyl-4-yl)-4-[(methoxyacetyl)amino]-L-arabinonic acid, Ubiquitin, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, Orlicky, S, Tyers, M, Sicheri, F. | | Deposit date: | 2013-08-22 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6095 Å) | | Cite: | E2 enzyme inhibition by stabilization of a low-affinity interface with ubiquitin.
Nat.Chem.Biol., 10, 2014
|
|
3MKS
 
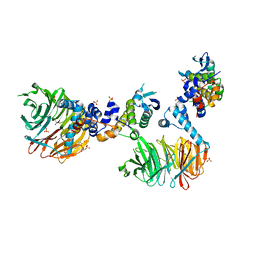 | | Crystal Structure of yeast Cdc4/Skp1 in complex with an allosteric inhibitor SCF-I2 | | Descriptor: | 1,1'-binaphthalene-2,2'-dicarboxylic acid, Cell division control protein 4, GLYCEROL, ... | | Authors: | Orlicky, S, Sicheri, F, Tyers, M, Tang, X. | | Deposit date: | 2010-04-15 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase.
Nat.Biotechnol., 28, 2010
|
|
4NKG
 
 | | Crystal structure of SspH1 LRR domain in complex PKN1 HR1b domain | | Descriptor: | E3 ubiquitin-protein ligase sspH1, HEXANE-1,6-DIOL, Serine/threonine-protein kinase N1 | | Authors: | Keszei, A.F.A, Xiaojing, T, Mccormick, C, Zeqiraj, E, Rohde, J.R, Tyers, M, Sicheri, F. | | Deposit date: | 2013-11-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase.
Mol.Cell.Biol., 34, 2014
|
|
4NKH
 
 | | Crystal structure of SspH1 LRR domain | | Descriptor: | E3 ubiquitin-protein ligase sspH1 | | Authors: | Keszei, A.F.A, Xiaojing, T, Mccormick, C, Zeqiraj, E, Rohde, J.R, Tyers, M, Sicheri, F. | | Deposit date: | 2013-11-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase.
Mol.Cell.Biol., 34, 2014
|
|
3V7D
 
 | | Crystal Structure of ScSkp1-ScCdc4-pSic1 peptide complex | | Descriptor: | Cell division control protein 4, Protein SIC1, Suppressor of kinetochore protein 1 | | Authors: | Tang, X, Orlicky, S, Mittag, T, Csizmok, V, Pawson, T, Forman-Kay, J, Sicheri, F, Tyers, M. | | Deposit date: | 2011-12-20 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Composite low affinity interactions dictate recognition of the cyclin-dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6OGN
 
 | | Crystal structure of mouse protein arginine methyltransferase 7 in complex with SGC8158 chemical probe | | Descriptor: | 5'-S-(4-{[(4'-chloro[1,1'-biphenyl]-3-yl)methyl]amino}butyl)-5'-thioadenosine, Protein arginine N-methyltransferase 7, UNKNOWN ATOM OR ION, ... | | Authors: | Halabelian, L, Dong, A, Zeng, H, Li, Y, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-03 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response.
Nat Commun, 11, 2020
|
|
4QLB
 
 | | Structural Basis for the Recruitment of Glycogen Synthase by Glycogenin | | Descriptor: | GLYCEROL, Probable glycogen [starch] synthase, Protein GYG-1, ... | | Authors: | Zeqiraj, E, Judd, A, Sicheri, F. | | Deposit date: | 2014-06-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recruitment of glycogen synthase by glycogenin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5IBK
 
 | | Skp1-F-box in complex with a ubiquitin variant | | Descriptor: | F-box/WD repeat-containing protein 7, Polyubiquitin-B, S-phase kinase-associated protein 1,S-phase kinase-associated protein 1 | | Authors: | Orlicky, S, Sicheri, F. | | Deposit date: | 2016-02-22 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Inhibition of SCF ubiquitin ligases by engineered ubiquitin variants that target the Cul1 binding site on the Skp1-F-box interface.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3I3T
 
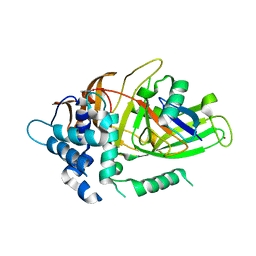 | | Crystal structure of covalent ubiquitin-USP21 complex | | Descriptor: | ETHANAMINE, Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 21, ... | | Authors: | Neculai, D, Avvakumov, G.V, Walker, J.R, Xue, S, Butler-Cole, C, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
4I6L
 
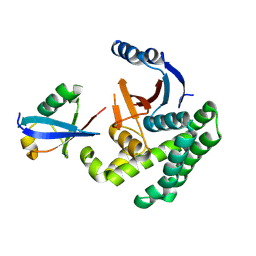 | |
2HJN
 
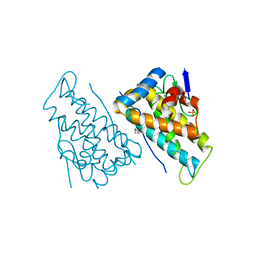 | |
3MTN
 
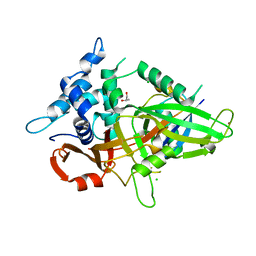 | | Usp21 in complex with a ubiquitin-based, USP21-specific inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, UBIQUITIN VARIANT UBV.21.4, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Ernst, A, Sidhu, S, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
3N3K
 
 | | The catalytic domain of USP8 in complex with a USP8 specific inhibitor | | Descriptor: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 8, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Allali-Hassani, A, Lam, R, Ernst, A, Sidhu, S, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
3BS7
 
 | |
3BS5
 
 | | Crystal Structure of hCNK2-SAM/dHYP-SAM Complex | | Descriptor: | Connector enhancer of kinase suppressor of ras 2, Protein aveugle | | Authors: | Rajakulendran, T, Ceccarelli, D.F, Kurinov, I, Sicheri, F. | | Deposit date: | 2007-12-22 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CNK and HYP form a discrete dimer by their SAM domains to mediate RAF kinase signaling.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3V6E
 
 | | Crystal Structure of USP2 and a mutant form of Ubiquitin | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin, ... | | Authors: | Neculai, M, Ernst, A, Sidhu, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
3RZ3
 
 | | Human Cdc34 E2 in complex with CC0651 inhibitor | | Descriptor: | 4,5-dideoxy-5-(3',5'-dichlorobiphenyl-4-yl)-4-[(methoxyacetyl)amino]-L-arabinonic acid, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, Webb, D.R, Sicheri, F. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An allosteric inhibitor of the human cdc34 ubiquitin conjugating enzyme
Cell(Cambridge,Mass.), 145, 2011
|
|
3BQ3
 
 | | Crystal structure of S. cerevisiae Dcn1 | | Descriptor: | Defective in cullin neddylation protein 1, GLYCEROL | | Authors: | Chou, Y.C, Sicheri, F. | | Deposit date: | 2007-12-19 | | Release date: | 2008-01-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dcn1 Functions as a Scaffold-Type E3 Ligase for Cullin Neddylation.
Mol.Cell, 29, 2008
|
|
7U3E
 
 | | GID4 in complex with compound 1 | | Descriptor: | Glucose-induced degradation protein 4 homolog, tert-butyl (1S,4S)-2,5-diazabicyclo[2.2.1]heptane-2-carboxylate | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3G
 
 | | GID4 in complex with compound 67 | | Descriptor: | (1R)-1-phenyl-1,2,3,4-tetrahydroisoquinolin-5-amine, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3I
 
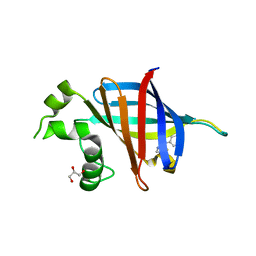 | | GID4 in complex with compound 16 | | Descriptor: | 3-{[(5S)-5-methyl-4,5-dihydro-1,3-thiazol-2-yl]amino}phenol, GLYCEROL, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3K
 
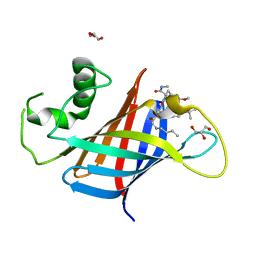 | | GID4 in complex with compound 89 | | Descriptor: | GLYCEROL, Glucose-induced degradation protein 4 homolog, N-butylglycyl-4-tert-butyl-D-phenylalanyl-3-methoxy-N-methyl-L-phenylalaninamide | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3F
 
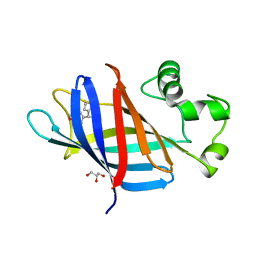 | | GID4 in complex with compound 4 | | Descriptor: | (4R)-4-(4-methoxyphenyl)-4,5,6,7-tetrahydrothieno[3,2-c]pyridine, GLYCEROL, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
