8RZW
 
 | | A fragment-based inhibitor of SHP2 | | Descriptor: | 3,5-bis(chloranyl)pyrazin-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Cleasby, A, Price, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
8S06
 
 | | A fragment-based inhibitor of SHP2 | | Descriptor: | 1H-pyrrolo[3,2-b]pyridin-7-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Cleasby, A, Price, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
8RZY
 
 | | A fragment-based inhibitor of SHP2 | | Descriptor: | 1H-pyrrolo[3,2-b]pyridin-5-amine, DIMETHYL SULFOXIDE, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Cleasby, A, Price, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
8S07
 
 | | A fragment-based inhibitor of SHP2 | | Descriptor: | 7-azanyl-N-pyridin-3-yl-3H-pyrrolo[3,2-b]pyridine-2-carboxamide, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Cleasby, A, Price, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
8S01
 
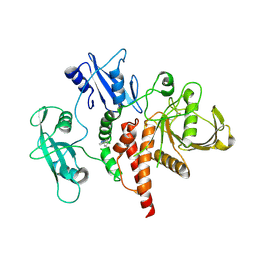 | | A fragment-based inhibitor of SHP2 | | Descriptor: | 3-(4-chlorophenyl)-1H-pyrazole, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Cleasby, A, Price, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
8S04
 
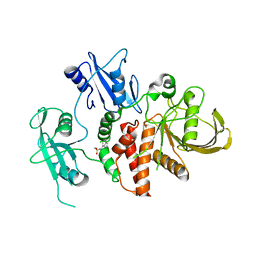 | | A fragment-based inhibitor of SHP2 | | Descriptor: | N-(1H-indol-7-yl)methanesulfonamide, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Cleasby, A, Price, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
8VZ9
 
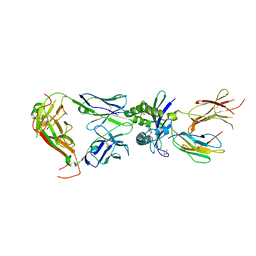 | | Crystal structure of mouse MAIT M2A TCR-MR1-5-OP-RU complex | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Ciacchi, L, Rossjohn, J, Awad, W. | | Deposit date: | 2024-02-11 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mouse mucosal-associated invariant T cell receptor recognition of MR1 presenting the vitamin B metabolite, 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil.
J.Biol.Chem., 2024
|
|
8VZ8
 
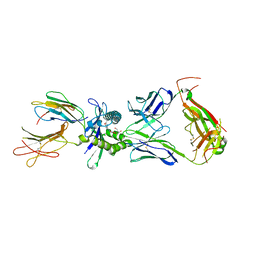 | | Crystal structure of mouse MAIT M2B TCR-MR1-5-OP-RU complex | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Ciacchi, L, Rossjohn, J, Awad, W. | | Deposit date: | 2024-02-11 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Mouse mucosal-associated invariant T cell receptor recognition of MR1 presenting the vitamin B metabolite, 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil.
J.Biol.Chem., 2024
|
|
8Y9X
 
 | | Crystal structure of the complex of lactoperoxidase with four inorganic substrates, SCN, I, Br and Cl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, CALCIUM ION, ... | | Authors: | Viswanathan, V, Singh, A.K, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for the order of preference of inorganic substrates in mammalian heme peroxidases: crystal structure of the complex of lactoperoxidase with four inorganic substrates, SCN, I, Br and Cl
To Be Published
|
|
8RXR
 
 | | Crystal structure of VPS34 in complex with inhibitor SB02024 | | Descriptor: | 4-[(3R)-3-methylmorpholin-4-yl]-2-[(2R)-2-(trifluoromethyl)piperidin-1-yl]-3H-pyridin-6-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Tresaugues, L, Yu, Y, Bogdan, M, Parpal, S, Silvander, C, Lindstrom, J, Simeon, J, Timson, M.J, Al-Hashimi, H, Smith, B.D, Flynn, D.L, Viklund, J, Martinsson, J, De Milito, A, Andersson, M. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Combining VPS34 inhibitors with STING agonists enhances type I interferon signaling and anti-tumor efficacy.
Mol Oncol, 2024
|
|
8RX9
 
 | | LTA4 hydrolase in complex with compound3 | | Descriptor: | 1-[[5-[5-(1~{H}-pyrazol-5-yl)pyridin-2-yl]oxypyridin-2-yl]methyl]piperidin-4-ol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2024-02-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Guided Elaboration of a Fragment-Like Hit into an Orally Efficacious Leukotriene A4 Hydrolase Inhibitor.
J.Med.Chem., 67, 2024
|
|
8RX3
 
 | | LTA4 hydrolase in complex with CTX-4430 | | Descriptor: | 4-[[(1~{S},4~{S})-5-[[4-[4-(1,3-oxazol-2-yl)phenoxy]phenyl]methyl]-2,5-diazabicyclo[2.2.1]heptan-2-yl]methyl]benzoic acid, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2024-02-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Elaboration of a Fragment-Like Hit into an Orally Efficacious Leukotriene A4 Hydrolase Inhibitor.
J.Med.Chem., 67, 2024
|
|
8RX7
 
 | | LTA4 hydrolase in complex with compound2 | | Descriptor: | 5-(4-phenoxyphenyl)-1~{H}-imidazole, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2024-02-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Elaboration of a Fragment-Like Hit into an Orally Efficacious Leukotriene A4 Hydrolase Inhibitor.
J.Med.Chem., 67, 2024
|
|
8Y6P
 
 | |
8Y6I
 
 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in nanodisc | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ATP-dependent translocase ABCB1,mNeonGreen, CHOLESTEROL, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
8Y6H
 
 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in LMNG detergent | | Descriptor: | ATP-dependent translocase ABCB1,mNeonGreen, UIC2 Fab heavy chain, UIC2 Fab light chain, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
8VW0
 
 | |
8VW2
 
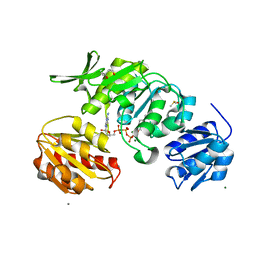 | |
8VW1
 
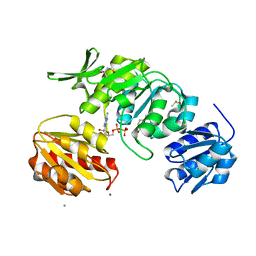 | |
8RV9
 
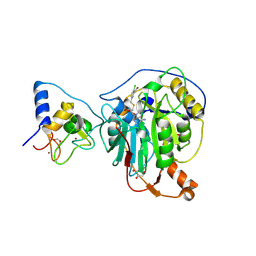 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 6 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-2-chloranyl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 6
To Be Published
|
|
8RV8
 
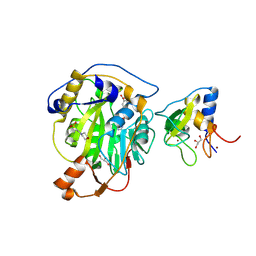 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 5 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]-2-chloranyl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 5
To Be Published
|
|
8RV6
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 2 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(4-hydroxyphenyl)benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 3
To Be Published
|
|
8RUF
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV D187A mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
8RUD
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV K138A mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
8RUG
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV C189A mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
