2RR1
 
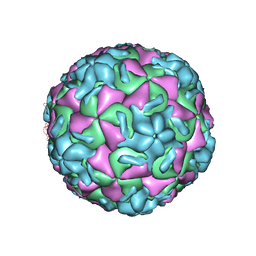 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
3INS
 
 | |
2LZ2
 
 | |
7DFR
 
 | |
6DFR
 
 | |
5DFR
 
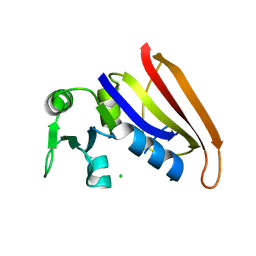 | |
1BDS
 
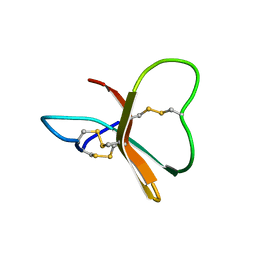 | |
2BDS
 
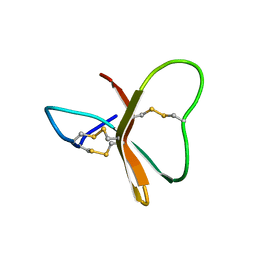 | |
1LLC
 
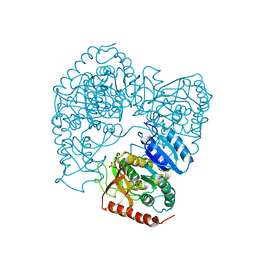 | | STRUCTURE DETERMINATION OF THE ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS CASEI AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | 1,6-di-O-phosphono-alpha-D-fructofuranose, L-LACTATE DEHYDROGENASE, SULFATE ION | | Authors: | Buehner, M, Hecht, H.J, Hensel, R. | | Deposit date: | 1988-11-21 | | Release date: | 1989-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | STRUCTURE DETERMINATION OF THE ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS-CASEI AT 3A RESOLUTION.
Acta Crystallogr.,Sect.A, 40, 1984
|
|
2CRO
 
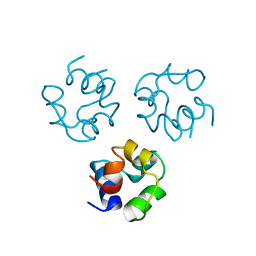 | |
1FD2
 
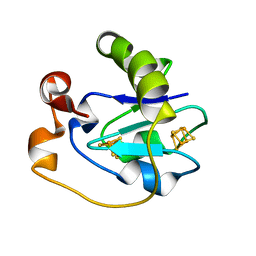 | |
1R69
 
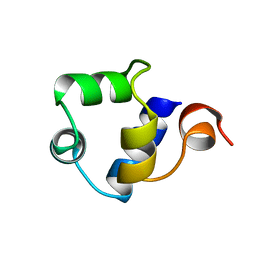 | | STRUCTURE OF THE AMINO-TERMINAL DOMAIN OF PHAGE 434 REPRESSOR AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | REPRESSOR PROTEIN CI | | Authors: | Mondragon, A, Subbiah, S, Alamo, S.C, Drottar, M, Harrison, S.C. | | Deposit date: | 1988-12-08 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the amino-terminal domain of phage 434 repressor at 2.0 A resolution.
J.Mol.Biol., 205, 1989
|
|
1DNF
 
 | | EFFECTS OF 5-FLUOROURACIL/GUANINE WOBBLE BASE PAIRS IN Z-DNA. MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGFG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(UFP)P*G)-3'), MAGNESIUM ION | | Authors: | Coll, M, Saal, D, Frederick, C.A, Aymami, J, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-12-12 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effects of 5-fluorouracil/guanine wobble base pairs in Z-DNA: molecular and crystal structure of d(CGCGFG).
Nucleic Acids Res., 17, 1989
|
|
1VTF
 
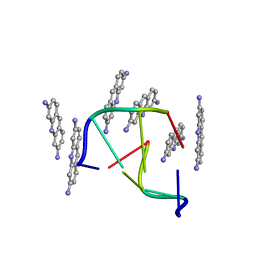 | |
2HIR
 
 | |
4HIR
 
 | |
1VTV
 
 | | Molecular structure of (M5DC-DG)3: The role of the methyl group on 5-methyl cytosine in stabilizing Z-DNA | | Descriptor: | DNA (5'-D(*(CH3)CP*GP*(CH3)CP*GP*(CH3)CP*G)-3') | | Authors: | Fujii, S, Wang, A.H.-J, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1989-01-10 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular Structure of (m5dC-dG)3: The Role of the Methyl Group on 5-Methyl Cytosine in Stabilizing Z-DNA
Nucleic Acids Res., 10, 1982
|
|
1LYD
 
 | |
1VJ4
 
 | | SEQUENCE-DEPENDENT CONFORMATION OF AN A-DNA DOUBLE HELIX: THE CRYSTAL STRUCTURE OF THE OCTAMER D(G-G-T-A-T-A-C-C) | | Descriptor: | 5'-D(*GP*GP*TP*AP*TP*AP*CP*C)-3' | | Authors: | Shakked, Z, Rabinovich, D, Kennard, O, Cruse, W.B, Salisbury, S.A, Viswamitra, M.A. | | Deposit date: | 1989-01-11 | | Release date: | 1989-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequence-dependent conformation of an A-DNA double helix: The crystal structure of the octamer d(G-G-T-A-T-A-C-C)
J.Mol.Biol., 166, 1983
|
|
1COH
 
 | |
1HOE
 
 | |
2CA2
 
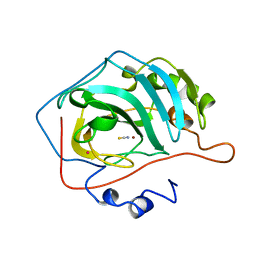 | | CRYSTALLOGRAPHIC STUDIES OF INHIBITOR BINDING SITES IN HUMAN CARBONIC ANHYDRASE II. A PENTACOORDINATED BINDING OF THE SCN-ION TO THE ZINC AT HIGH P*H | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, THIOCYANATE ION, ... | | Authors: | Eriksson, A.E, Kylsten, P.M, Jones, T.A, Liljas, A. | | Deposit date: | 1989-02-06 | | Release date: | 1990-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic studies of inhibitor binding sites in human carbonic anhydrase II: a pentacoordinated binding of the SCN- ion to the zinc at high pH.
Proteins, 4, 1988
|
|
1CA2
 
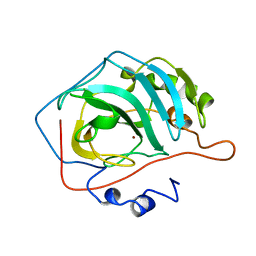 | |
1MBA
 
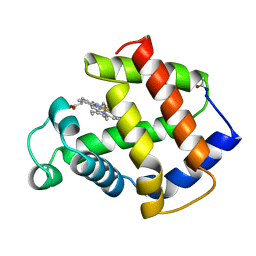 | | APLYSIA LIMACINA MYOGLOBIN. CRYSTALLOGRAPHIC ANALYSIS AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Onesti, S, Gatti, G, Coda, A, Ascenzi, P, Brunori, M. | | Deposit date: | 1989-02-22 | | Release date: | 1990-01-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aplysia limacina myoglobin. Crystallographic analysis at 1.6 A resolution.
J.Mol.Biol., 205, 1989
|
|
3MBA
 
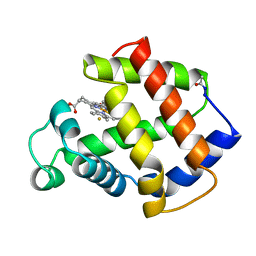 | | APLYSIA LIMACINA MYOGLOBIN. CRYSTALLOGRAPHIC ANALYSIS AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | FLUORIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Onesti, S, Gatti, G, Coda, A, Ascenzi, P, Brunori, M. | | Deposit date: | 1989-02-22 | | Release date: | 1990-01-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aplysia limacina myoglobin. Crystallographic analysis at 1.6 A resolution.
J.Mol.Biol., 205, 1989
|
|
