6I0I
 
 | |
6HNL
 
 | | Selenomethionine derivative of IdmH 96-104 loop truncation variant | | Descriptor: | Putative polyketide cyclase IdmH | | Authors: | Drulyte, I, Obajdin, J, Trinh, C, Hemsworth, G.R, Berry, A. | | Deposit date: | 2018-09-16 | | Release date: | 2019-11-06 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the putative cyclase IdmH from the indanomycin nonribosomal peptide synthase/polyketide synthase.
Iucrj, 6, 2019
|
|
4L9U
 
 | | Structure of C-terminal coiled coil of RasGRP1 | | Descriptor: | GLYCEROL, RAS guanyl-releasing protein 1, SULFATE ION | | Authors: | Iwig, J.S, Vercoulen, Y, Das, R, Barros, T, Limnander, A, Che, Y, Pelton, J.G, Wemmer, D.E, Roose, J.P, Kuriyan, J. | | Deposit date: | 2013-06-18 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6014 Å) | | Cite: | Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1.
Elife, 2, 2013
|
|
4L9M
 
 | | Autoinhibited state of the Ras-specific exchange factor RasGRP1 | | Descriptor: | CITRIC ACID, GLYCEROL, RAS guanyl-releasing protein 1, ... | | Authors: | Iwig, J.S, Vercoulen, Y, Das, R, Barros, T, Limnander, A, Che, Y, Pelton, J.G, Wemmer, D.E, Roose, J.P, Kuriyan, J. | | Deposit date: | 2013-06-18 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1.
Elife, 2, 2013
|
|
4LCD
 
 | |
4L6V
 
 | | Crystal structure of a virus like photosystem I from the cyanobacterium Synechocystis PCC 6803 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Mazor, Y, Nataf, D, Toporik, H, Nelson, N. | | Deposit date: | 2013-06-13 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structures of virus-like photosystem I complexes from the mesophilic cyanobacterium Synechocystis PCC 6803.
Elife, 3, 2014
|
|
1I5T
 
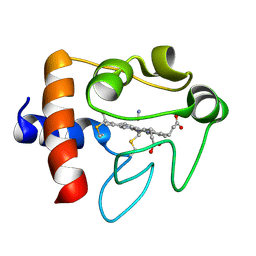 | | SOLUTION STRUCTURE OF CYANOFERRICYTOCHROME C | | Descriptor: | CYANIDE ION, CYTOCHROME C, HEME C | | Authors: | Yao, Y, Qian, C, Ye, K, Wang, J, Tang, W. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cyanoferricytochrome c: ligand-controlled conformational flexibility and electronic structure of the heme moiety.
J.Biol.Inorg.Chem., 7, 2002
|
|
4M8M
 
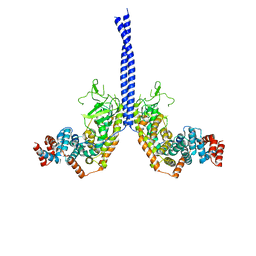 | |
4MEX
 
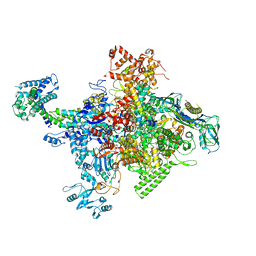 | | Crystal structure of Escherichia coli RNA polymerase in complex with salinamide A | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Feng, Y, Zhang, Y, Arnold, E, Ebright, R.H. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-21 | | Last modified: | 2014-06-25 | | Method: | X-RAY DIFFRACTION (3.902 Å) | | Cite: | Transcription inhibition by the depsipeptide antibiotic salinamide A.
Elife, 3, 2014
|
|
4M8T
 
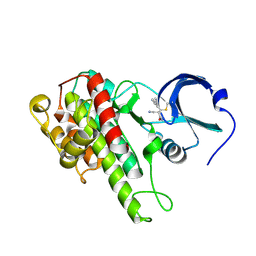 | |
4M8N
 
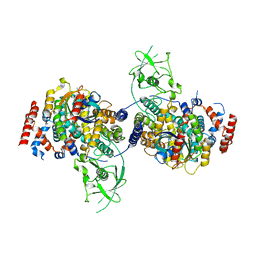 | | Crystal Structure of PlexinC1/Rap1B Complex | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pascoe, H.G, Wang, Y, Brautigam, C.A, He, H, Zhang, X. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structural basis for activation and non-canonical catalysis of the Rap GTPase activating protein domain of plexin.
Elife, 2, 2013
|
|
4MEY
 
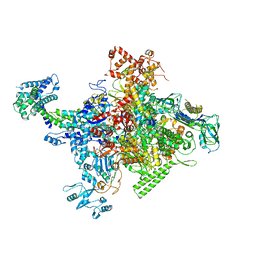 | | Crystal structure of Escherichia coli RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Feng, Y, Zhang, Y, Arnold, E, Ebright, R.H. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.948 Å) | | Cite: | Transcription inhibition by the depsipeptide antibiotic salinamide A.
Elife, 3, 2014
|
|
4MQ9
 
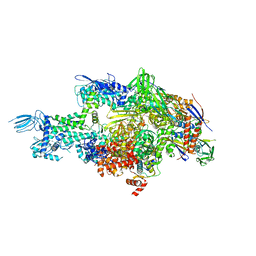 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme in complex with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ho, M.X, Arnold, E, Ebright, R.H, Zhang, Y, Tuske, S. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
1K0T
 
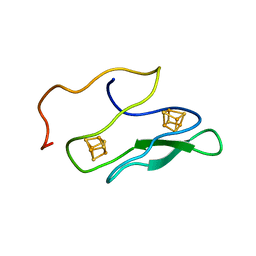 | | NMR SOLUTION STRUCTURE OF UNBOUND, OXIDIZED PHOTOSYSTEM I SUBUNIT PSAC, CONTAINING [4FE-4S] CLUSTERS FA AND FB | | Descriptor: | IRON/SULFUR CLUSTER, PSAC SUBUNIT OF PHOTOSYSTEM I | | Authors: | Antonkine, M.L, Liu, G, Bentrop, D, Bryant, D.A, Bertini, I, Luchinat, C, Golbeck, J.H, Stehlik, D. | | Deposit date: | 2001-09-20 | | Release date: | 2002-06-05 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the unbound, oxidized Photosystem I subunit PsaC, containing [4Fe-4S] clusters F(A) and F(B): a conformational change occurs upon binding to photosystem I.
J.Biol.Inorg.Chem., 7, 2002
|
|
4NEE
 
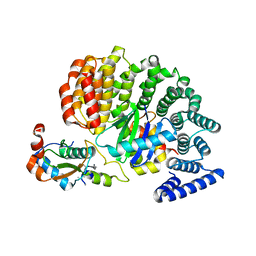 | | crystal structure of AP-2 alpha/simga2 complex bound to HIV-1 Nef | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit sigma, Protein Nef | | Authors: | Hurley, J.H, Bonifacino, J.S, Ren, X, Park, S.Y. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8841 Å) | | Cite: | How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4.
Elife, 3, 2014
|
|
4NM0
 
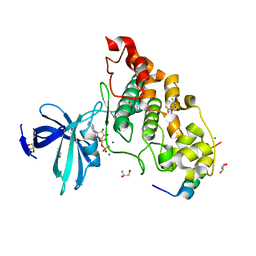 | | Crystal structure of peptide inhibitor-free GSK-3/Axin complex | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE-5'-DIPHOSPHATE, Axin-1, ... | | Authors: | Chu, M.L.-H, Stamos, J.L, Enos, M.D, Shah, N, Weis, W.I. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of GSK-3 inhibition by N-terminal phosphorylation and by the Wnt receptor LRP6.
Elife, 3, 2014
|
|
3J4G
 
 | |
3J4U
 
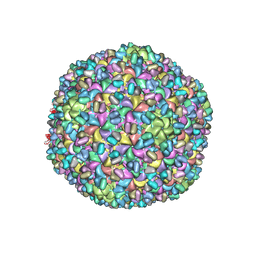 | | A new topology of the HK97-like fold revealed in Bordetella bacteriophage: non-covalent chainmail secured by jellyrolls | | Descriptor: | cementing protein, major capsid protein | | Authors: | Zhang, X, Guo, H, Jin, L, Czornyj, E, Hodes, A, Hui, W.H, Nieh, A.W, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 A resolution.
Elife, 2, 2013
|
|
3J6C
 
 | | Cryo-EM structure of MAVS CARD filament | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | Xu, H, He, X, Zheng, H, Huang, L.J, Hou, F, Yu, Z, de la Cruz, M.J, Borkowski, B, Zhang, X, Chen, Z.J, Jiang, Q.-X. | | Deposit date: | 2014-02-04 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Structural basis for the prion-like MAVS filaments in antiviral innate immunity.
Elife, 3, 2014
|
|
3J2Z
 
 | |
6LCE
 
 | |
3J2X
 
 | |
3J30
 
 | |
4OIP
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077, ATP, and CMPcPP | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
3J4Q
 
 | |
