8SMI
 
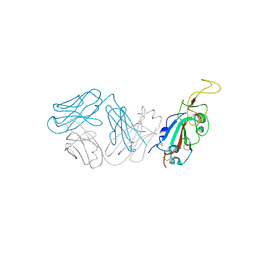 | | Crystal structure of antibody WRAIR-2123 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, WRAIR-2123 Fab heavy chain, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
8FG2
 
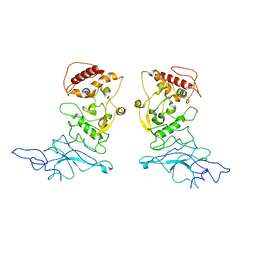 | | SARS-CoV-2 Nucleocapsid dimer structure determined from COVID-19 patients | | Descriptor: | Nucleoprotein | | Authors: | Casasanta, M, Jonaid, G.M, Kaylor, L, Luqiu, W, DiCecco, L, Solares, M, Berry, S, Kelly, D.F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural Insights of the SARS-CoV-2 Nucleocapsid Protein: Implications for the Inner-workings of Rapid Antigen Tests.
Microsc Microanal, 29, 2023
|
|
8U1G
 
 | |
8TYJ
 
 | |
7C8V
 
 | | Structure of sybody SR4 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7C8W
 
 | | Structure of sybody MR17 in complex with the SARS-CoV-2 S receptor-binding domain (RBD) | | Descriptor: | GLYCEROL, Spike protein S1, Synthetic nanobody MR17, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7CAN
 
 | | Structure of sybody MR17-K99Y in complex with the SARS-CoV-2 S Receptor-binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-09 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
8GIA
 
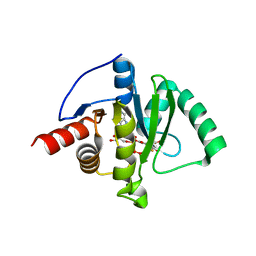 | | Crystal structure of SARS-CoV-2 (Covid-19) Nsp3 macrodomain in complex with TFMU-ADPr | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl [(2R,3S,4R,5R)-3,4-dihydroxy-5-{[2-oxo-4-(trifluoromethyl)-2H-1-benzopyran-7-yl]oxy}oxolan-2-yl]methyl dihydrogen diphosphate | | Authors: | Wallace, S.D, Bagde, S.R, Fromme, J.C. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Fluorescence Polarization Assay for Macrodomains Facilitates the Identification of Potent Inhibitors of the SARS-CoV-2 Macrodomain.
Acs Chem.Biol., 18, 2023
|
|
7URS
 
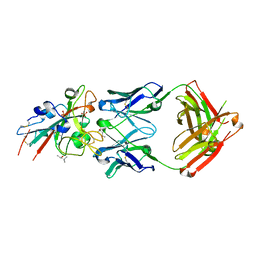 | |
7URQ
 
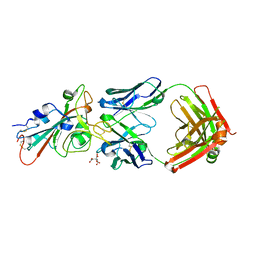 | |
7VMU
 
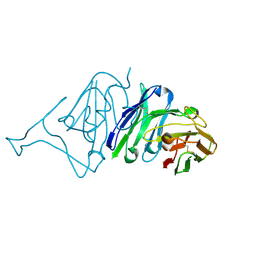 | | Crystal Structure of SARS-CoV Spike Receptor-Binding Domain Complexed with Neutralizing Antibody | | Descriptor: | Spike protein S1, scFv E4 | | Authors: | Guo, Y, Wang, W, Jiao, P, Yang, H, Rao, Z, Cheng, G. | | Deposit date: | 2021-10-09 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Antibody engineering improves neutralization activity against K417 spike mutant SARS-CoV-2 variants.
Cell Biosci, 12, 2022
|
|
7R7N
 
 | |
7RN0
 
 | |
7RN1
 
 | |
7RXD
 
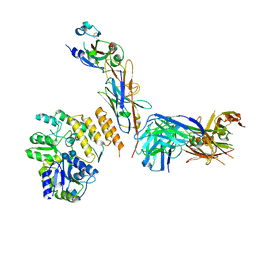 | | CryoEM structure of RBD domain of COVID-19 in complex with Legobody | | Descriptor: | Fab_8D3_2 heavy chain, Fab_8D3_2 light chain, Maltodextrin-binding protein,Immunoglobulin G-binding protein A,Immunoglobulin G-binding protein G, ... | | Authors: | Wu, X.D, Rapoport, T.A. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure determination of small proteins by nanobody-binding scaffolds (Legobodies).
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S4S
 
 | |
7KX5
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with noncovalent inhibitor Jun8-76-3A | | Descriptor: | 3C-like proteinase, GLYCEROL, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Di- and Trihaloacetamides as Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity.
J.Am.Chem.Soc., 143, 2021
|
|
7KN4
 
 | |
7KN3
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain complexed with a pre-pandemic antibody S-B8 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, S-B8 Fab heavy chain, ... | | Authors: | Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago.
Adv Sci, 9, 2022
|
|
7LYI
 
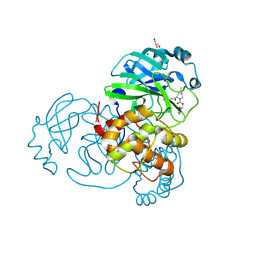 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-3 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7M6H
 
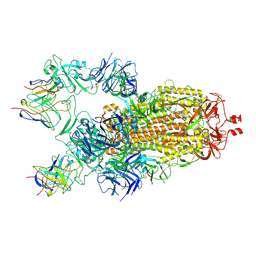 | |
7M6D
 
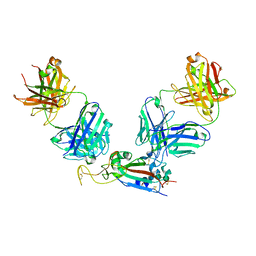 | |
7M6G
 
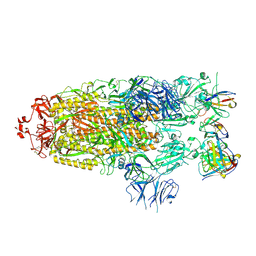 | |
7M6I
 
 | |
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
