6FK9
 
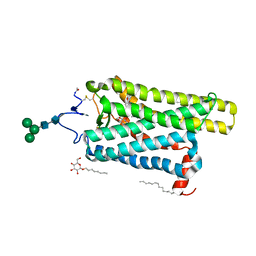 | | Crystal structure of N2C/D282C stabilized opsin bound to RS09 | | Descriptor: | (2~{S})-3-methyl-2-phenyl-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-butan-1-one, PALMITIC ACID, Rhodopsin, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4Z7B
 
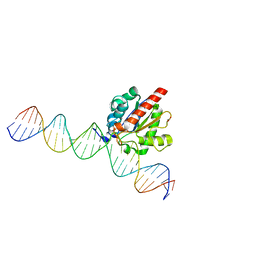 | | Structure of the enzyme-product complex resulting from TDG action on a GfC mismatch | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DNA (28-MER), ... | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-04-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
6FKB
 
 | | Crystal structure of N2C/D282C stabilized opsin bound to RS13 | | Descriptor: | 2-(4-chlorophenyl)-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FK8
 
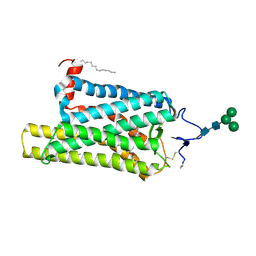 | | Crystal structure of N2C/D282C stabilized opsin bound to RS08 | | Descriptor: | (2~{R},3~{S})-3-azanyl-2-(4-chlorophenyl)-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-butan-1-one, PALMITIC ACID, Rhodopsin, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FKD
 
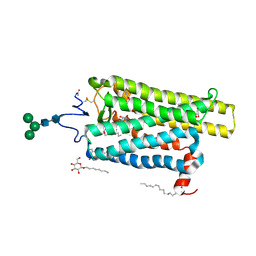 | | Crystal structure of N2C/D282C stabilized opsin bound to RS16 | | Descriptor: | 5-chloranyl-2-(2-oxidanylidene-2-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-ethyl)-3~{H}-pyridin-6-one, PALMITIC ACID, Rhodopsin, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FK7
 
 | | Crystal structure of N2C/D282C stabilized opsin bound to RS06 | | Descriptor: | (2~{R},3~{R})-2-(4-chlorophenyl)-3-oxidanyl-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-butan-1-one, PALMITIC ACID, Rhodopsin, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FPL
 
 | | TETR(D) E147A MUTANT IN COMPLEX WITH TETRACYCLINE AND MAGNESIUM | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, TETRACYCLINE, ... | | Authors: | Hinrichs, W, Palm, G.J, Berndt, L, Girbardt, B. | | Deposit date: | 2018-02-11 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
1MS7
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-Des-Me-AMPA at 1.97 A resolution, Crystallization in the presence of zinc acetate | | Descriptor: | (S)-2-AMINO-3-(3-HYDROXY-ISOXAZOL-4-YL)PROPIONIC ACID, Glutamate receptor subunit 2, ZINC ION | | Authors: | Kasper, C, Lunn, M.-L, Liljefors, T, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2002-09-19 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | GluR2 ligand-binding core complexes: importance of the isoxazolol moiety and 5-substituent for the binding mode of AMPA-type agonists
FEBS Lett., 531, 2002
|
|
1N0T
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with the antagonist (S)-ATPO at 2.1 A resolution. | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Greenwood, J.R, Liljefors, T, Lunn, M.-L, Egebjerg, J, Larsen, I.K, Gouaux, E, Kastrup, J.S. | | Deposit date: | 2002-10-15 | | Release date: | 2003-03-04 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Competitive antagonism of AMPA receptors by ligands of
different classes: crystal structure of ATPO bound to the
GluR2 ligand-binding core, in comparison with DNQX.
J.Med.Chem., 46, 2003
|
|
1MZT
 
 | |
1NNP
 
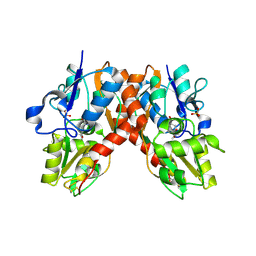 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-ATPA at 1.9 A resolution. Crystallization without zinc ions. | | Descriptor: | 3-(5-TERT-BUTYL-3-OXIDOISOXAZOL-4-YL)-L-ALANINATE, Glutamate receptor 2, SULFATE ION | | Authors: | Lunn, M.L, Hogner, A, Stensbol, T.B, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2003-01-14 | | Release date: | 2003-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Structure of the Ligand-Binding
Core of GluR2 in Complex with the Agonist (S)-ATPA:
Implications for Receptor Subunit Selectivity.
J.Med.Chem., 46, 2003
|
|
1NNK
 
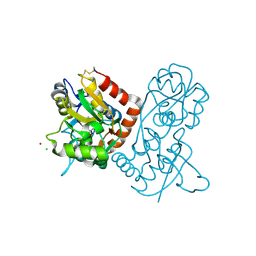 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-ATPA at 1.85 A resolution. Crystallization with zinc ions. | | Descriptor: | 3-(5-TERT-BUTYL-3-OXIDOISOXAZOL-4-YL)-L-ALANINATE, CHLORIDE ION, Glutamate receptor 2, ... | | Authors: | Lunn, M.-L, Hogner, A, Stensbol, T.B, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2003-01-14 | | Release date: | 2003-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of the Ligand-Binding
Core of GluR2 in Complex with the Agonist (S)-ATPA:
Implications for Receptor Subunit Selectivity.
J.Med.Chem., 46, 2003
|
|
4LAL
 
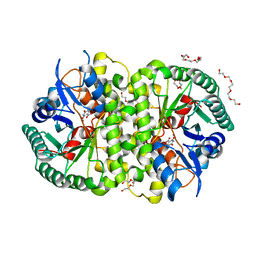 | | Crystal structure of Cordyceps militaris IDCase D323A mutant in complex with 5-carboxyl-uracil | | Descriptor: | 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, HEXAETHYLENE GLYCOL, Uracil-5-carboxylate decarboxylase, ... | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4LAM
 
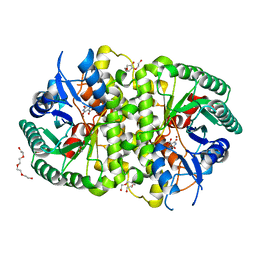 | | Crystal structure of Cordyceps militaris IDCase D323N mutant in complex with 5-carboxyl-uracil | | Descriptor: | 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, HEXAETHYLENE GLYCOL, Uracil-5-carboxylate decarboxylase, ... | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4LAN
 
 | | Crystal structure of Cordyceps militaris IDCase H195A mutant | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4LAO
 
 | | Crystal structure of Cordyceps militaris IDCase H195A mutant (Zn) | | Descriptor: | Cordyceps militaris IDCase, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4LAK
 
 | | Crystal structure of Cordyceps militaris IDCase D323N mutant in apo form | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Li, W, Zhu, J, Wang, R, Li, Z, Xu, G.L, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4MC2
 
 | | HIV protease in complex with SA525P | | Descriptor: | (3S)-tetrahydrofuran-3-yl {(2S,3R)-4-[(4R)-4-tert-butyl-7-fluoro-1,1-dioxido-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, Protease | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
4MC6
 
 | | HIV protease in complex with SA499 | | Descriptor: | 1,2-ETHANEDIOL, 1-tert-butyl-3-{(2S,3R)-4-[(4R)-7-fluoro-1,1-dioxido-4-(propan-2-yl)-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}urea, CHLORIDE ION, ... | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Antropow, A, White, A, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
4MCD
 
 | | hinTrmD in complex with 5-PHENYLTHIENO[2,3-D]PYRIMIDIN-4(3H)-ONE | | Descriptor: | 5-phenylthieno[2,3-d]pyrimidin-4(3H)-one, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Lahiri, S. | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selective Inhibitors of Bacterial t-RNA-(N(1)G37) Methyltransferase (TrmD) That Demonstrate Novel Ordering of the Lid Domain.
J.Med.Chem., 56, 2013
|
|
5DYS
 
 | | Crystal Structure of T94I rhodopsin mutant | | Descriptor: | ACETATE ION, PALMITIC ACID, RETINAL, ... | | Authors: | Singhal, A, Guo, Y, Matkovic, M, Schertler, G, Deupi, X, Yan, E, Standfuss, J. | | Deposit date: | 2015-09-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural role of the T94I rhodopsin mutation in congenital stationary night blindness.
Embo Rep., 17, 2016
|
|
4MC1
 
 | | HIV protease in complex with SA526P | | Descriptor: | (3S)-tetrahydrofuran-3-yl {(2S,3R)-4-[(4S)-4-tert-butyl-7-fluoro-1,1-dioxido-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, Protease | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Antropow, A, White, A, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
4MC9
 
 | | HIV protease in complex with AA74 | | Descriptor: | (3S)-tetrahydrofuran-3-yl {(2S,3R)-4-[(4R)-7-fluoro-1,1-dioxido-4-(propan-2-yl)-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Antropow, A, White, A, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
5EN0
 
 | | Crystal Structure of T94I rhodopsin mutant | | Descriptor: | ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-3, PALMITIC ACID, ... | | Authors: | Singhal, A, Guo, Y, Matkovic, M, Schertler, G, Deupi, X, Yan, E, Standfuss, J. | | Deposit date: | 2015-11-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural role of the T94I rhodopsin mutation in congenital stationary night blindness.
Embo Rep., 17, 2016
|
|
5EXH
 
 | |
