2XPV
 
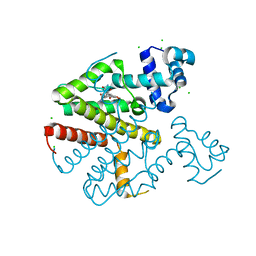 | | TetR(D) in complex with minocycline and magnesium. | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Dalm, D, Proft, J, Palm, G.J, Hinrichs, W. | | Deposit date: | 2010-08-30 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Tetr(D) in Complex with Minocycline.
To be Published
|
|
8HD6
 
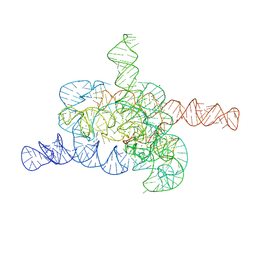 | | The relaxed pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The relaxed pre-Tet-S1 state molecule of co-transcriptional folded G264A mutant Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
8HD7
 
 | | The intermediate pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The intermediate pre-Tet-S1 state molecule of co-transcriptional folded G264A mutant Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
8I7N
 
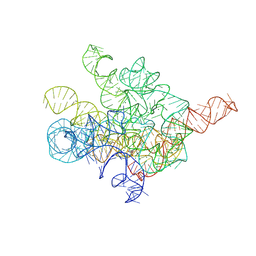 | | The Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Descriptor: | (2R,3R,4S,5R)-2-(2-azanylpurin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD3
 
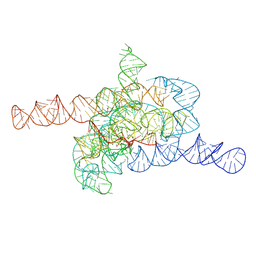 | | The relaxed pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, The relaxed pre-Tet-S1 state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
3R4A
 
 | |
3R4H
 
 | |
8A3I
 
 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil homotetramer with 3 heptad repeats, apCC-Tet*3 | | Descriptor: | apCC-Tet*3 | | Authors: | Naudin, E.A, Mylemans, B, Albanese, K.I, Woolfson, D.N. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | From peptides to proteins: coiled-coil tetramers to single-chain 4-helix bundles.
Chem Sci, 13, 2022
|
|
8A3G
 
 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil homotetramer with 4 heptad repeats, apCC-Tet* | | Descriptor: | ACETATE ION, SODIUM ION, apCC-Tet* | | Authors: | Naudin, E.A, Mylemans, B, Albanese, K.I, Woolfson, D.N. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | From peptides to proteins: coiled-coil tetramers to single-chain 4-helix bundles.
Chem Sci, 13, 2022
|
|
5TUE
 
 | | Crystal structure of tetracycline destructase Tet(50) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Tetracycline destructase Tet(50) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUI
 
 | | Crystal structure of tetracycline destructase Tet(50) in complex with chlortetracycline | | Descriptor: | 7-CHLOROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUK
 
 | | Crystal structure of tetracycline destructase Tet(51) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Tetracycline destructase Tet(51) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUM
 
 | | Crystal structure of tetracycline destructase Tet(56) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, tetracycline destructase Tet(56) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUL
 
 | |
5TUF
 
 | | Crystal structure of tetracycline destructase Tet(50) in complex with anhydrotetracycline | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
2XPU
 
 | | TetR(D) in complex with anhydrotetracycline. | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, CHLORIDE ION, SULFATE ION, ... | | Authors: | Dalm, D, Palm, G.J, Hinrichs, W. | | Deposit date: | 2010-08-30 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Tetracycline Repressor Allostery Does not Depend on Divalent Metal Recognition.
Biochemistry, 53, 2014
|
|
8HMA
 
 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with tetrandrine (TET) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6,6',7,12-tetramethoxy-2,2'-dimethyl-1beta-3,4-didehydroberbaman, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
7XD4
 
 | | The intermediate pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Descriptor: | Co-transcriptional folded wild-type Tetrahymena group I intron with 6nt 3'/5'-exon, MAGNESIUM ION | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
6F3K
 
 | | Combined solid-state NMR, solution-state NMR and EM data for structure determination of the tetrahedral aminopeptidase TET2 from P. horikoshii | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Gauto, D.F, Estrozi, L.F, Schwieters, C.D, Effantin, G, Macek, P, Sounier, R, Kerfah, R, Sivertsen, A.C, Colletier, J.P, Boisbouvier, J, Schoehn, G, Favier, A, Schanda, P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-03-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLID-STATE NMR, SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
1M5C
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH Br-HIBO AT 1.65 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5E
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH ACPA AT 1.46 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
8F8M
 
 | | LRH-1 bound to small molecule Tet and fragment of coactivator Tif2 | | Descriptor: | (1~{R},3~{a}~{R},6~{a}~{R})-4-phenyl-3~{a}-(1-phenylethenyl)-5-[9-(1~{H}-1,2,3,4-tetrazol-5-yl)nonyl]-2,3,6,6~{a}-tetrahydro-1~{H}-pentalen-1-ol, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-11-22 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Isosteric improvements to liver receptor homolog-1 small molecule modulators
To Be Published
|
|
6R8N
 
 | | STRUCTURE DETERMINATION OF THE TETRAHEDRAL AMINOPEPTIDASE TET2 FROM P. HORIKOSHII BY USE OF COMBINED SOLID-STATE NMR, SOLUTION-STATE NMR AND EM DATA 4.1 A, FOLLOWED BY REAL_SPACE_REFINEMENT AT 4.1 A | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Colletier, J.-P, Gauto, D, Estrozi, L, Favier, A, Effantin, G, Schoehn, G, Boisbouvier, J, Schanda, P. | | Deposit date: | 2019-04-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
1A2N
 
 | | STRUCTURE OF THE C115A MUTANT OF MURA COMPLEXED WITH THE FLUORINATED ANALOG OF THE REACTION TETRAHEDRAL INTERMEDIATE | | Descriptor: | UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL-3-FLUORO-2-PHOSPHONOOXY)PROPIONIC ACID | | Authors: | Skarzynski, T. | | Deposit date: | 1998-01-06 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Stereochemical course of enzymatic enolpyruvyl transfer and catalytic conformation of the active site revealed by the crystal structure of the fluorinated analogue of the reaction tetrahedral intermediate bound to the active site of the C115A mutant of MurA
Biochemistry, 37, 1998
|
|
4Y0P
 
 | | Bovine beta-lactoglobulin complex with tetracaine (BLG-TET) | | Descriptor: | Beta-lactoglobulin, Tetracaine | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Jablonski, M, Czub, M, Ye, X, Lewinski, K. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | beta-Lactoglobulin interactions with local anaesthetic drugs - Crystallographic and calorimetric studies.
Int.J.Biol.Macromol., 80, 2015
|
|
