8VJJ
 
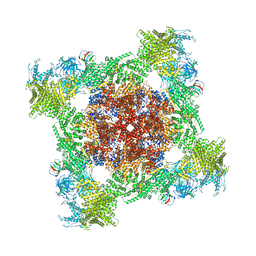 | | Structure of mouse RyR1 (EGTA-only dataset) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ... | | Authors: | Weninger, G, Marks, A.R. | | Deposit date: | 2024-01-07 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural insights into the regulation of RyR1 by S100A1.
To Be Published
|
|
8VIB
 
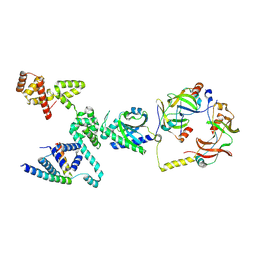 | |
8VID
 
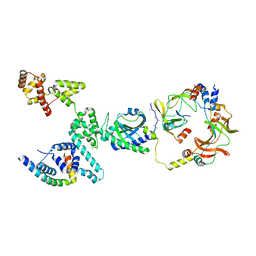 | |
8VHM
 
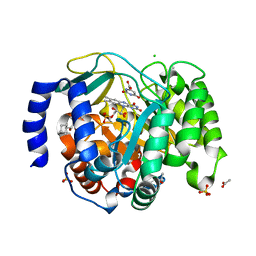 | | Structure of DHODH in Complex with Fragment 2 | | Descriptor: | ACETATE ION, CHLORIDE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of Alternative Binding Poses through Fragment-Based Identification of DHODH Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8VHL
 
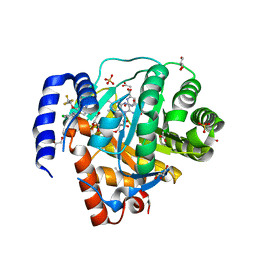 | | Structure of DHODH in Complex with Ligand 17 | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of Alternative Binding Poses through Fragment-Based Identification of DHODH Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8VGZ
 
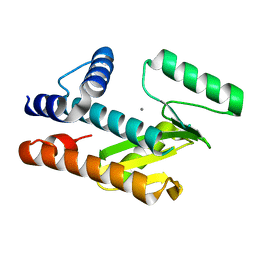 | |
8RKS
 
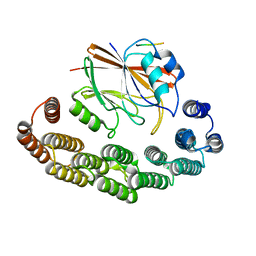 | | Structure of VPS29-VPS35 bound to the LFa motif R21 of Fam21. | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35, WASH complex subunit 2A | | Authors: | Romano-Moreno, M, Astorga-Simon, E.N, Rojas, A.L, Hierro, A. | | Deposit date: | 2023-12-30 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Retromer-mediated recruitment of the WASH complex involves discrete interactions between VPS35, VPS29, and FAM21.
Protein Sci., 33, 2024
|
|
8VGC
 
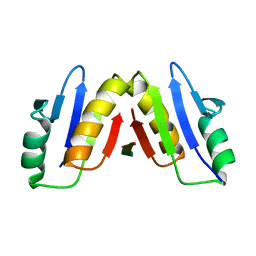 | | Complex of ExbD with D-box peptide: Orthorhombic form | | Descriptor: | Biopolymer transport protein ExbD, GLN-PRO-ILE-SER-VAL-THR-MET-VAL-THR-PRO | | Authors: | Loll, P.J. | | Deposit date: | 2023-12-27 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery and structural characterization of the D-box, a conserved TonB motif that couples an inner-membrane motor to outer-membrane transport.
J.Biol.Chem., 300, 2024
|
|
8VGD
 
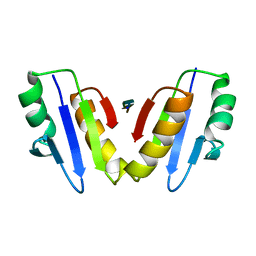 | | Complex of ExbD with D-box peptide: Tetragonal form | | Descriptor: | Biopolymer transport protein ExbD, GLN-PRO-ILE-SER-VAL-THR-MET-VAL-THR | | Authors: | Loll, P.J. | | Deposit date: | 2023-12-27 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery and structural characterization of the D-box, a conserved TonB motif that couples an inner-membrane motor to outer-membrane transport.
J.Biol.Chem., 300, 2024
|
|
8RKI
 
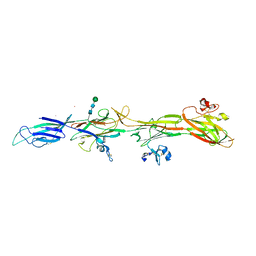 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament fragment | | Descriptor: | Choriogenin H, YTTERBIUM (III) ION, Zona pellucida sperm-binding protein 3, ... | | Authors: | Wiseman, B, Zamora-Caballero, S, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8RK2
 
 | |
8VEV
 
 | |
8RJC
 
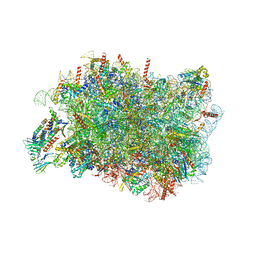 | |
8RJD
 
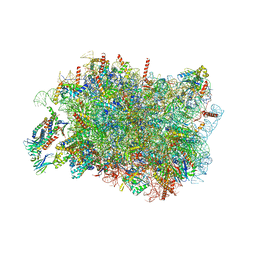 | |
8RJB
 
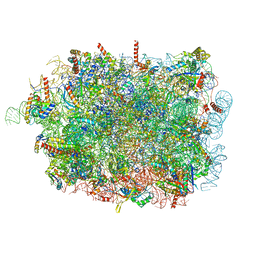 | |
8XI6
 
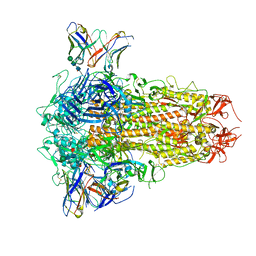 | | SARS-CoV-2 Omicron BQ.1.1 Variant Spike Protein Complexed with MO11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ishimaru, H, Nishimura, M, Shigematsu, H, Marini, M.I, Hasegawa, N, Takamiya, R, Iwata, S, Mori, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Epitopes of an antibody that neutralizes a wide range of SARS-CoV-2 variants in a conserved subdomain 1 of the spike protein.
J.Virol., 2024
|
|
8RIV
 
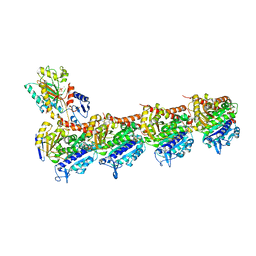 | | T2R-TTL-1-K08 complex | | Descriptor: | (4-fluoranyl-2-methyl-1~{H}-indol-5-yl) 3,4,5-trimethoxybenzenesulfonate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Boiarska, Z, Homer, J.A, Steinmetz, M.O, Moses, J.E, Prota, A.E.P. | | Deposit date: | 2023-12-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Modular synthesis of functional libraries by accelerated SuFEx click chemistry.
Chem Sci, 15, 2024
|
|
8RIW
 
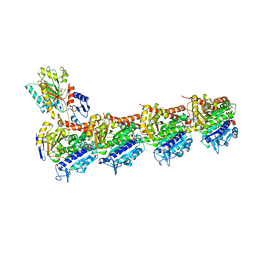 | | T2R-TTL-1-L01 complex | | Descriptor: | (2-methyl-1~{H}-indol-5-yl) 3,4,5-trimethoxybenzenesulfonate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E.P, Boiarska, Z, Homer, J.A, Steinmetz, M.O, Moses, J.E. | | Deposit date: | 2023-12-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Modular synthesis of functional libraries by accelerated SuFEx click chemistry.
Chem Sci, 15, 2024
|
|
8RI0
 
 | |
8RHR
 
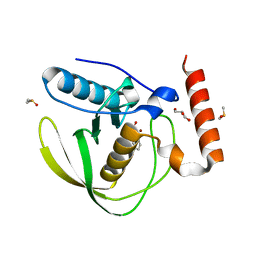 | | E.coli Peptide Deformylase with bound inhibitor BB4 | | Descriptor: | 2-(5-bromo-1H-indol-3-yl)-N-hydroxyacetamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Kirschner, H, Stoll, R, Hofmann, E. | | Deposit date: | 2023-12-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Toward More Selective Antibiotic Inhibitors: A Structural View of the Complexed Binding Pocket of E. coli Peptide Deformylase.
J.Med.Chem., 2024
|
|
8XGC
 
 | | Structure of yeast replisome associated with FACT and histone hexamer, Composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Li, N, Gao, Y, Yu, D, Gao, N, Zhai, Y. | | Deposit date: | 2023-12-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Parental histone transfer caught at the replication fork.
Nature, 627, 2024
|
|
8RHI
 
 | | Lytic Transglycosylase MltD of Pseudomonas aeruginosa in a ternary complex bound to Bulgecin A and chito-tetraose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-(4-O-SULFONYL-N-ACETYLGLUCOSAMININYL)-5-METHYLHYDROXY-L-PROLINE-TAURINE, ... | | Authors: | Miguel-Ruano, V, Hermoso, J.A. | | Deposit date: | 2023-12-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural characterization of lytic transglycosylase MltD of Pseudomonas aeruginosa, a target for the natural product bulgecin A.
Int.J.Biol.Macromol., 267, 2024
|
|
8RHE
 
 | |
8RHF
 
 | | Lytic Transglycosylase MltD of Pseudomonas aeruginosa bound to the Natural Product Bulgecin A, with two LysM domains | | Descriptor: | 4-O-(4-O-SULFONYL-N-ACETYLGLUCOSAMININYL)-5-METHYLHYDROXY-L-PROLINE-TAURINE, DI(HYDROXYETHYL)ETHER, LysM peptidoglycan-binding domain-containing protein, ... | | Authors: | Miguel-Ruano, V, Hermoso, J.A. | | Deposit date: | 2023-12-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of lytic transglycosylase MltD of Pseudomonas aeruginosa, a target for the natural product bulgecin A.
Int.J.Biol.Macromol., 267, 2024
|
|
8VCI
 
 | | SARS-CoV-2 Frameshift Stimulatory Element with Upstream Multibranch Loop | | Descriptor: | Frameshift Stimulatory Element with Upstream Multi-branch Loop | | Authors: | Peterson, J.M, Becker, S.T, O'Leary, C.A, Juneja, P, Yang, Y, Moss, W.N. | | Deposit date: | 2023-12-14 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | SARS-CoV-2 Frameshift Stimulatory Element with Upstream Multibranch Loop
To Be Published
|
|
