3O1B
 
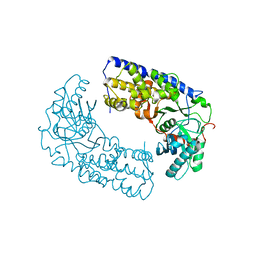 | | CRYSTAL STRUCTURE OF DIMERIC KLHXK1 IN CRYSTAL FORM II | | Descriptor: | Hexokinase | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-21 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O1W
 
 | | Crystal structure of dimeric KlHxk1 in crystal form III | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Hexokinase, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-22 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O4W
 
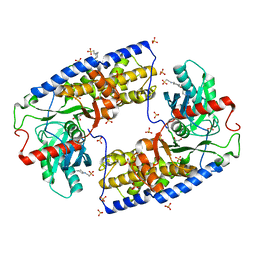 | | Crystal structure of dimeric KlHxk1 in crystal form IV | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Hexokinase, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-27 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O5B
 
 | | Crystal structure of dimeric KlHxk1 in crystal form VII with glucose bound (open state) | | Descriptor: | Hexokinase, SULFATE ION, beta-D-glucopyranose | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O6W
 
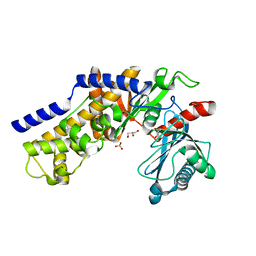 | | Crystal structure of monomeric KlHxk1 in crystal form VIII (open state) | | Descriptor: | GLYCEROL, Hexokinase, PHOSPHATE ION | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-29 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O80
 
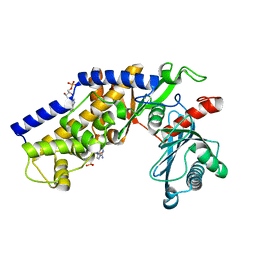 | | Crystal structure of monomeric KlHxk1 in crystal form IX (open state) | | Descriptor: | Hexokinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O8M
 
 | | Crystal structure of monomeric KlHxk1 in crystal form XI with glucose bound (closed state) | | Descriptor: | CHLORIDE ION, Hexokinase, alpha-D-glucopyranose, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3QIC
 
 | | The structure of human glucokinase E339K mutation | | Descriptor: | GLYCEROL, Glucokinase, alpha-D-glucopyranose | | Authors: | Liu, Q, Liu, S, Liu, J. | | Deposit date: | 2011-01-27 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of E339K mutated human glucokinase reveals changes in the ATP binding site.
Febs Lett., 585, 2011
|
|
3S41
 
 | | Glucokinase in complex with activator and glucose | | Descriptor: | Glucokinase, N,N-dimethyl-5-({2-methyl-6-[(5-methylpyrazin-2-yl)carbamoyl]-1-benzofuran-4-yl}oxy)pyrimidine-2-carboxamide, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2011-05-18 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Designing glucokinase activators with reduced hypoglycemia risk: discovery of N,N-dimethyl-5-(2-methyl-6-((5-methylpyrazin-2-yl)-carbamoyl)benzofuran-4-yloxy)pyrimidine-2-carboxamide as a clinical candidate for the treatment of type 2 diabetes mellitus
MEDCHEMCOMM, 2, 2011
|
|
3VEV
 
 | | Glucokinase in complex with an activator and glucose | | Descriptor: | (2S)-3-cyclopentyl-N-(5-methylpyridin-2-yl)-2-[2-oxo-4-(trifluoromethyl)pyridin-1(2H)-yl]propanamide, Glucokinase, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-09 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
3VEY
 
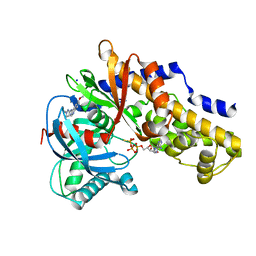 | | glucokinase in complex with glucose and ATPgS | | Descriptor: | 6-methoxy-N-(1-methyl-1H-pyrazol-3-yl)quinazolin-4-amine, Glucokinase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-09 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
3VF6
 
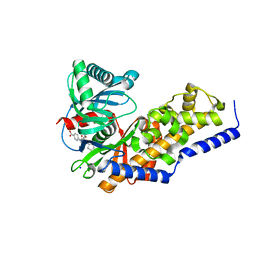 | | Glucokinase in complex with glucose and activator | | Descriptor: | 6-({(2S)-3-cyclopentyl-2-[4-(trifluoromethyl)-1H-imidazol-1-yl]propanoyl}amino)pyridine-3-carboxylic acid, Glucokinase, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-09 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
3W0L
 
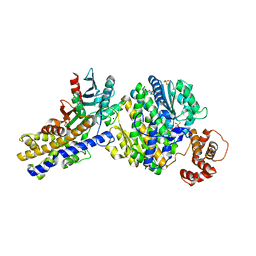 | | The crystal structure of Xenopus Glucokinase and Glucokinase Regulatory Protein complex | | Descriptor: | FRUCTOSE -6-PHOSPHATE, Glucokinase, Glucokinase regulatory protein, ... | | Authors: | Choi, J.M, Seo, M.H, Kyeong, H.H, Kim, E, Kim, H.S. | | Deposit date: | 2012-10-31 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Molecular basis for the role of glucokinase regulatory protein as the allosteric switch for glucokinase
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4DCH
 
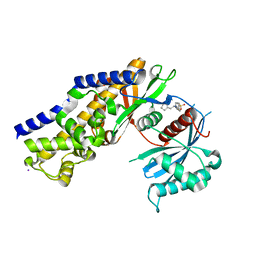 | | Insights into Glucokinase Activation Mechanism: Observation of Multiple Distinct Protein Conformations | | Descriptor: | (2R)-3-cyclopentyl-2-[4-(methylsulfonyl)phenyl]-N-(1,3-thiazol-2-yl)propanamide, Glucokinase, IODIDE ION, ... | | Authors: | Greasley, S.E, Hickey, M, Feng, J, Garcia, E. | | Deposit date: | 2012-01-17 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
4DHY
 
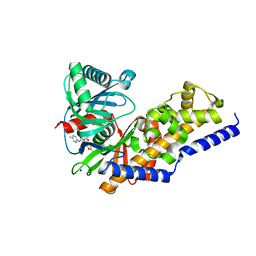 | | Crystal structure of human glucokinase in complex with glucose and activator | | Descriptor: | Glucokinase, N,N-dimethyl-5-({2-methyl-6-[(5-methylpyrazin-2-yl)carbamoyl]-1-benzofuran-4-yl}oxy)pyrimidine-2-carboxamide, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
4F9O
 
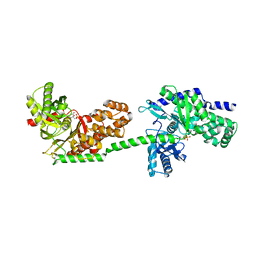 | |
4FOE
 
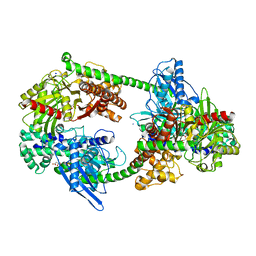 | |
4FOI
 
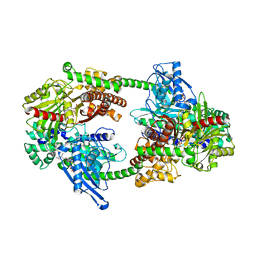 | |
4FPA
 
 | |
4FPB
 
 | |
4ISE
 
 | | Human glucokinase in complex with novel activator (2S)-3-cyclohexyl-2-(6-fluoro-4-oxoquinazolin-3(4H)-yl)-N-(1,3-thiazol-2-yl)propanamide | | Descriptor: | (2S)-3-cyclohexyl-2-(6-fluoro-4-oxoquinazolin-3(4H)-yl)-N-(1,3-thiazol-2-yl)propanamide, Glucokinase, IODIDE ION, ... | | Authors: | Hosfield, D, Skene, R.J. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Design, synthesis and SAR of novel glucokinase activators.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4ISF
 
 | | Human glucokinase in complex with novel activator (2S)-3-cyclohexyl-2-(6-fluoro-2,4-dioxo-1,4-dihydroquinazolin-3(2H)-yl)-N-(1,3-thiazol-2-yl)propanamide | | Descriptor: | (2S)-3-cyclohexyl-2-(6-fluoro-2,4-dioxo-1,4-dihydroquinazolin-3(2H)-yl)-N-(1,3-thiazol-2-yl)propanamide, Glucokinase, IODIDE ION, ... | | Authors: | Hosfield, D, Skene, R.J. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Design, synthesis and SAR of novel glucokinase activators.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4ISG
 
 | | Human glucokinase in complex with novel activator (2S)-3-cyclohexyl-2-[4-(methylsulfonyl)-2-oxopiperazin-1-yl]-N-(1,3-thiazol-2-yl)propanamide | | Descriptor: | (2S)-3-cyclohexyl-2-[4-(methylsulfonyl)-2-oxopiperazin-1-yl]-N-(1,3-thiazol-2-yl)propanamide, Glucokinase, IODIDE ION, ... | | Authors: | Hosfield, D, Skene, R.J. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Design, synthesis and SAR of novel glucokinase activators.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4IWV
 
 | | Crystals structure of Human Glucokinase in complex with small molecule activator | | Descriptor: | (2S)-2-{[1-(2-chlorophenyl)-1H-pyrazolo[3,4-d]pyrimidin-4-yl]oxy}-N-(5-chloropyridin-2-yl)-3-(2-hydroxyethoxy)propanamide, Glucokinase isoform 3, SODIUM ION, ... | | Authors: | Ogg, D.J, Hargreaves, D, Gerhardt, S, Flavell, L, McAlister, M. | | Deposit date: | 2013-01-24 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimising pharmacokinetics of glucokinase activators with matched triplicate design sets the discovery of AZD3651 and AZD9485
To be Published
|
|
4IXC
 
 | |
