7JFY
 
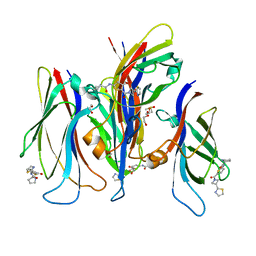 | | GAS41 YEATS domain in complex with 5 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, N-(5-{3-[(2S)-1,3-thiazolidin-2-yl]azetidine-1-carbonyl}thiophen-2-yl)-L-prolinamide, ... | | Authors: | Linhares, B.M, Listunov, D, Winkler, A, Grembecka, J, Cierpicki, T. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.100557 Å) | | Cite: | GAS41 YEATS domain in complex with 5
To Be Published
|
|
7F4A
 
 | |
7F5M
 
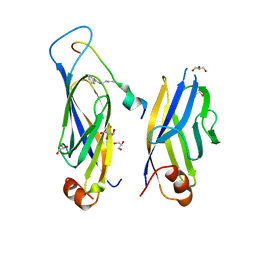 | |
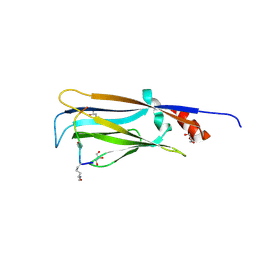
7EIC
 
 | |
7EID
 
 | |
7E7C
 
 | |
7EIE
 
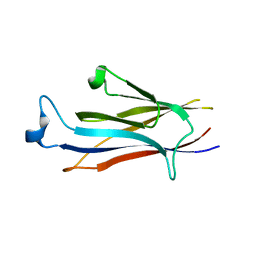 | | Crystal structure of YEATS2 YEATS domain | | Descriptor: | GLYCEROL, YEATS domain-containing protein 2 | | Authors: | Kikuchi, M, Umehara, T. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Elucidation of binding preferences of YEATS domains to site-specific acetylated nucleosome core particles.
J.Biol.Chem., 298, 2022
|
|
7EIF
 
 | | Crystal structure of GAS41 YEATS domain | | Descriptor: | GLYCEROL, YEATS domain-containing protein 4 | | Authors: | Kikuchi, M, Umehara, T. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | GAS41 promotes H2A.Z deposition through recognition of the N terminus of histone H3 by the YEATS domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7VKH
 
 | |
7VKG
 
 | |
8DKB
 
 | |
7X8G
 
 | |
7X8B
 
 | |
7X8F
 
 | |
7X88
 
 | |
8I60
 
 | |
8IIY
 
 | |
8IIZ
 
 | |
8IJ0
 
 | |
8PJI
 
 | | MLLT1 in complex with compound 10a | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Protein ENL, ... | | Authors: | Raux, B, Diaz-Saez, L, Huber, K.V.M, Fedorov, O, Owen, D.R, Londregan, A.T, Bountra, C, Edwards, A, Arrowsmith, C. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of PFI-6, a small-molecule chemical probe for the YEATS domain of MLLT1 and MLLT3.
Bioorg.Med.Chem.Lett., 98, 2023
|
|
8PJ7
 
 | | MLLT3 in complex with compound PFI-6 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Raux, B, Diaz-Saez, L, Huber, K.V.M, Fedorov, O, Owen, D.R, Londregan, A.T, Bountra, C, Edwards, A, Arrowsmith, C. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery of PFI-6, a small-molecule chemical probe for the YEATS domain of MLLT1 and MLLT3.
Bioorg.Med.Chem.Lett., 98, 2023
|
|
8X1C
 
 | | Structure of nucleosome-bound SRCAP-C in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of nucleosome-bound SRCAP-C in the ADP-bound state
To Be Published
|
|
8X19
 
 | | Structure of nucleosome-bound SRCAP-C in the ADP-BeFx-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of nucleosome-bound SRCAP-C in the ADP-BeFx-bound state
To Be Published
|
|
8X15
 
 | | Structure of nucleosome-bound SRCAP-C in the apo state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of nucleosome-bound SRCAP-C in the apo state
To Be Published
|
|
