4CNS
 
 | | Crystal structure of truncated human CRMP-4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Crmp-4: Correction of Intensities for Lattice-Translocation Disorder
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CNT
 
 | | CRYSTAL STRUCTURE OF WT HUMAN CRMP-4 | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROPYRIMIDINASE-LIKE 3, SODIUM ION | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of Human Crmp-4: Correction of Intensities for Lattice-Translocation Disorder
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4DYK
 
 | | Crystal structure of an adenosine deaminase from pseudomonas aeruginosa pao1 (target nysgrc-200449) with bound zn | | Descriptor: | AMIDOHYDROLASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Raushel, F.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an adenosine deaminase from pseudomonas aeruginosa pao1 (target nysgrc-200449) with bound zn
to be published
|
|
4DZH
 
 | | Crystal structure of an adenosine deaminase from xanthomonas campestris (target nysgrc-200456) with bound zn | | Descriptor: | AMIDOHYDROLASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Raushel, F.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Crystal structure of an adenosine deaminase from xanthomonas campestris (target nysgrc-200456) with bound zn
to be published
|
|
4F0R
 
 | | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) bound Zn and 5'-Methylthioadenosine (unproductive complex) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5-methylthioadenosine/S-adenosylhomocysteine deaminase, GLYCEROL, ... | | Authors: | Kim, J, Vetting, M.W, Sauder, J.M, Burley, S.K, Raushel, F.M, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-04 | | Release date: | 2012-06-06 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) bound Zn and 5'-Methylthioadenosine (unproductive complex)
To be Published
|
|
4F0L
 
 | |
4F0S
 
 | | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) with bound inosine. | | Descriptor: | 5-methylthioadenosine/S-adenosylhomocysteine deaminase, CHLORIDE ION, INOSINE, ... | | Authors: | Kim, J, Vetting, M.W, Sauder, J.M, Burley, S.K, Raushel, F.M, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-04 | | Release date: | 2012-06-06 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) with bound inosine.
To be Published
|
|
4GBD
 
 | | Crystal Structure Of Adenosine Deaminase From Pseudomonas Aeruginosa Pao1 with bound Zn and methylthio-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, PHOSPHATE ION, Putative uncharacterized protein, ... | | Authors: | Ho, M, Guan, R, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Methylthioadenosine deaminase in an alternative quorum sensing pathway in Pseudomonas aeruginosa.
Biochemistry, 51, 2012
|
|
4H00
 
 | | The crystal structure of mon-Zn dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structural Requirements for Post-translational Modification
To be Published
|
|
4GZ7
 
 | | The crystal structure of Apo-dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, dihydropyrimidinase | | Authors: | Hsien, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structure Requirements for Post-translational Modification
To be Published
|
|
4H01
 
 | | The crystal structure of di-Zn dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structural Requirements for Post-translational Modification
To be Published
|
|
3NQB
 
 | |
3T81
 
 | |
3T8L
 
 | | Crystal Structure of adenine deaminase with Mn/Fe | | Descriptor: | Adenine deaminase 2, UNKNOWN ATOM OR ION | | Authors: | Bagaria, A, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-08-01 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The catalase activity of diiron adenine deaminase.
Protein Sci., 20, 2011
|
|
8A18
 
 | | 1.63 A resolution hydroquinone inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Ciurli, S, Cianci, M. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Inhibition of Urease by Hydroquinones: A Structural and Kinetic Study.
Chemistry, 28, 2022
|
|
5OL4
 
 | | 1.28 A resolution of Sporosarcina pasteurii urease inhibited in the presence of NBPT | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Phosphoramidothioic O,O-acid, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2017-07-26 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Urease Inhibition in the Presence of N-(n-Butyl)thiophosphoric Triamide, a Suicide Substrate: Structure and Kinetics.
Biochemistry, 56, 2017
|
|
4UBP
 
 | | STRUCTURE OF BACILLUS PASTEURII UREASE INHIBITED WITH ACETOHYDROXAMIC ACID AT 1.55 A RESOLUTION | | Descriptor: | ACETOHYDROXAMIC ACID, NICKEL (II) ION, PROTEIN (UREASE (CHAIN A)), ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 1999-02-25 | | Release date: | 2000-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The complex of Bacillus pasteurii urease with acetohydroxamate anion from X-ray data at 1.55 A resolution.
J.Biol.Inorg.Chem., 5, 2000
|
|
7P7O
 
 | | X-RAY CRYSTAL STRUCTURE OF SPOROSARCINA PASTEURII UREASE INHIBITED BY THE GOLD(I)-DIPHOSPHINE COMPOUND Au(PEt3)2Cl DETERMINED AT 1.87 ANGSTROMS | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, OXYGEN ATOM, ... | | Authors: | Mazzei, L, Ciurli, S, Cianci, M, Messori, L, Massai, L. | | Deposit date: | 2021-07-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Medicinal Au(I) compounds targeting urease as prospective antimicrobial agents: unveiling the structural basis for enzyme inhibition.
Dalton Trans, 50, 2021
|
|
7P7N
 
 | |
6YL3
 
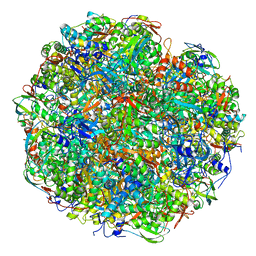 | | High resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica | | Descriptor: | NICKEL (II) ION, Urease subunit alpha, Urease subunit beta, ... | | Authors: | Righetto, R.D, Anton, L, Adaixo, R, Jakob, R, Zivanov, J, Mahi, M.A, Ringler, P, Schwede, T, Maier, T, Stahlberg, H. | | Deposit date: | 2020-04-06 | | Release date: | 2020-05-06 | | Last modified: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | High-resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica.
Nat Commun, 11, 2020
|
|
6ZJA
 
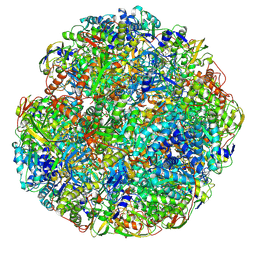 | | Helicobacter pylori urease with inhibitor bound in the active site | | Descriptor: | 2-{[1-(3,5-dimethylphenyl)-1H-imidazol-2-yl]sulfanyl}-N-hydroxyacetamide, NICKEL (II) ION, Urease subunit alpha, ... | | Authors: | Luecke, H, Cunha, E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-12-23 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 angstrom resolution.
Nat Commun, 12, 2021
|
|
4Z42
 
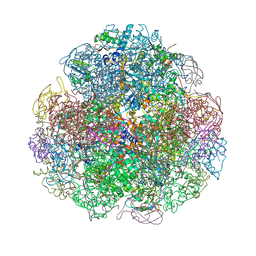 | | Crystal structure of urease from Yersinia enterocolitica | | Descriptor: | NICKEL (II) ION, Urease subunit alpha, Urease subunit beta, ... | | Authors: | Studer, G, Jakob, R.P, Mahi, M.A, Wiesand, U, Schwede, T, Maier, T. | | Deposit date: | 2015-04-01 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure of urease from Yersinia enterocolitica
To Be Published
|
|
5A6T
 
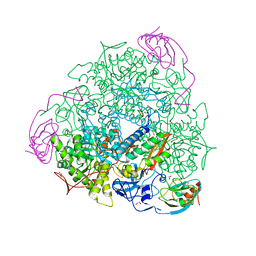 | | 1.65 A resolution Sulphite inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Mazzei, L, Cianci, M, Benini, S, Bertini, L, Musiani, F, Ciurli, S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic and Structural Studies Reveal a Unique Binding Mode of Sulfite to the Nickel Center in Urease.
J.Inorg.Biochem., 154, 2015
|
|
6G48
 
 | | Sporosarcina pasteurii urease inhibited by silver | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2018-03-27 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The structure of urease inactivated by Ag(i): a new paradigm for enzyme inhibition by heavy metals.
Dalton Trans, 47, 2018
|
|
7ZCY
 
 | | Sporosarcina pasteurii urease (SPU) co-crystallized in the presence of an Ebselen-derivative and bound to Se atoms | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, N-(2-chloranyl-4-fluoranyl-phenyl)-2-selanyl-benzamide, ... | | Authors: | Mazzei, L, Ciurli, S, Cianci, M. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Optimized Ebselen-Based Inhibitors of Bacterial Ureases with Nontypical Mode of Action.
J.Med.Chem., 66, 2023
|
|
