4V6M
 
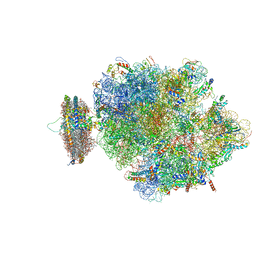 | | Structure of the ribosome-SecYE complex in the membrane environment | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 16S RIBOSOMAL RNA, ... | | Authors: | Frauenfeld, J, Gumbart, J, van der Sluis, E.O, Funes, S, Gartmann, M, Beatrix, B, Mielke, T, Berninghausen, O, Becker, T, Schulten, K, Beckmann, R. | | Deposit date: | 2011-02-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Cryo-EM structure of the ribosome-SecYE complex in the membrane environment.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3S84
 
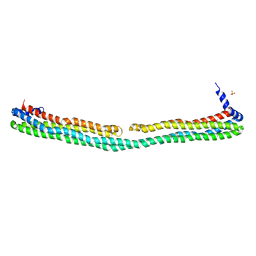 | | Dimeric apoA-IV | | Descriptor: | Apolipoprotein A-IV, SULFATE ION | | Authors: | Deng, X, Davidson, W.S, Thompson, T.B. | | Deposit date: | 2011-05-27 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of Dimeric Apolipoprotein A-IV and Its Mechanism of Self-Association.
Structure, 20, 2012
|
|
3R2P
 
 | |
3K2S
 
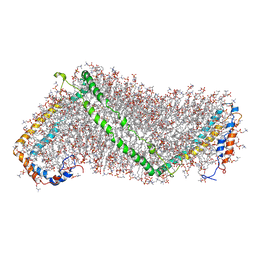 | | Solution structure of double super helix model | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Apolipoprotein A-I, CHOLESTEROL | | Authors: | Wu, Z, Gogonea, V, Lee, X, Wagner, M.A, Li, X.-M, Huang, Y, Undurti, A, May, R.P, Haertlein, M, Moulin, M, Gutsche, I, Zaccai, G, Didonato, J.A, Hazen, L.S. | | Deposit date: | 2009-09-30 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Double superhelix model of high density lipoprotein.
J.Biol.Chem., 284, 2009
|
|
2N5E
 
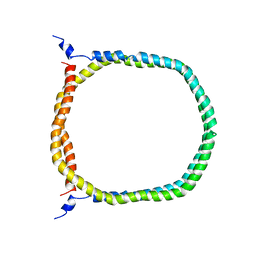 | | The 3D solution structure of discoidal high-density lipoprotein particles | | Descriptor: | Apolipoprotein A-I | | Authors: | Bibow, S, Polyhach, Y, Eichmann, C, Chi, C.N, Kowal, J, Stahlberg, H, Jeschke, G, Guentert, P, Riek, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-12-21 | | Last modified: | 2017-02-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of discoidal high-density lipoprotein particles with a shortened apolipoprotein A-I.
Nat.Struct.Mol.Biol., 24, 2017
|
|
2MSE
 
 | | NMR data-driven model of GTPase KRas-GNP:ARafRBD complex tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSD
 
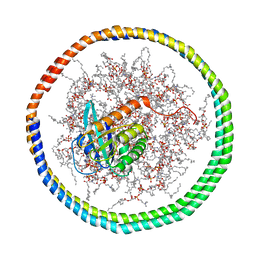 | | NMR data-driven model of GTPase KRas-GNP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSC
 
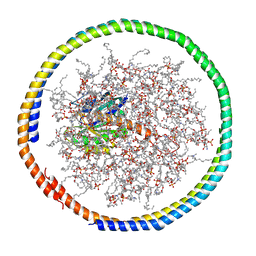 | | NMR data-driven model of GTPase KRas-GDP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2LEM
 
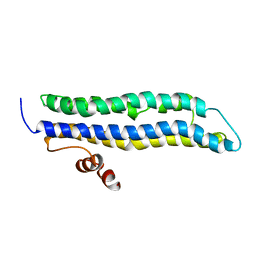 | |
2L7B
 
 | | NMR Structure of full length apoE3 | | Descriptor: | Apolipoprotein E | | Authors: | Chen, J, Wang, J. | | Deposit date: | 2010-12-07 | | Release date: | 2011-08-03 | | Last modified: | 2011-09-21 | | Method: | SOLUTION NMR | | Cite: | Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2KC3
 
 | |
1YA9
 
 | | Crystal Structure of the 22kDa N-Terminal Fragment of Mouse Apolipoprotein E | | Descriptor: | Apolipoprotein E | | Authors: | Peters-Libeu, C.A, Rutenber, E, Newhouse, Y, Hatters, D.M, Weisgraber, K.H. | | Deposit date: | 2004-12-17 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Engineering conformational destabilization into mouse apolipoprotein E. A model for a unique property of human apolipoprotein E4
J.Biol.Chem., 280, 2005
|
|
1OR3
 
 | | APOLIPOPROTEIN E3 (APOE3), TRIGONAL TRUNCATION MUTANT 165 | | Descriptor: | PROTEIN (APOLIPOPROTEIN E) | | Authors: | Rupp, B, Segelke, B.W. | | Deposit date: | 1998-12-01 | | Release date: | 2000-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Conformational flexibility in the apolipoprotein E amino-terminal domain structure determined from three new crystal forms: implications for lipid binding.
Protein Sci., 9, 2000
|
|
1OR2
 
 | | APOLIPOPROTEIN E3 (APOE3) TRUNCATION MUTANT 165 | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Rupp, B, Segelke, B.W, Forstner, M. | | Deposit date: | 1999-03-25 | | Release date: | 2000-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational flexibility in the apolipoprotein E amino-terminal domain structure determined from three new crystal forms: implications for lipid binding.
Protein Sci., 9, 2000
|
|
1NFO
 
 | | APOLIPOPROTEIN E2 (APOE2, D154A MUTATION) | | Descriptor: | APOLIPOPROTEIN E2 | | Authors: | Rupp, B, Parkin, S. | | Deposit date: | 1996-07-17 | | Release date: | 1997-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia.
Nat.Struct.Biol., 3, 1996
|
|
1NFN
 
 | | APOLIPOPROTEIN E3 (APOE3) | | Descriptor: | APOLIPOPROTEIN E3 | | Authors: | Rupp, B, Parkin, S. | | Deposit date: | 1996-07-17 | | Release date: | 1997-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia.
Nat.Struct.Biol., 3, 1996
|
|
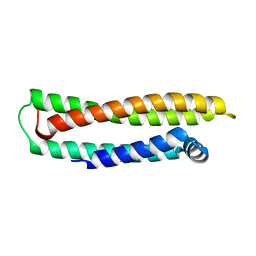
1LPE
 
 | |
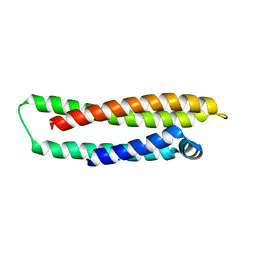
1LE4
 
 | |
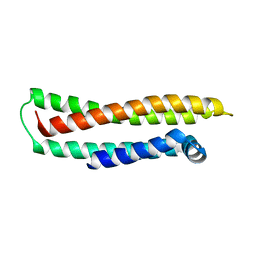
1LE2
 
 | |
1H7I
 
 | |
1GS9
 
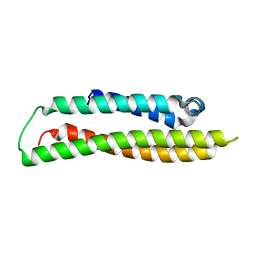 | | Apolipoprotein E4, 22k domain | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Verderame, J.R, Kantardjieff, K, Segelke, B, Weisgraber, K, Rupp, B. | | Deposit date: | 2002-01-02 | | Release date: | 2003-06-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the 22K Domain of Human Apolipoprotein E4
To be Published
|
|
1EA8
 
 | |
1BZ4
 
 | | APOLIPOPROTEIN E3 (APO-E3), TRUNCATION MUTANT 165 | | Descriptor: | PROTEIN (APOLIPOPROTEIN E) | | Authors: | Rupp, B, Segelke, B. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational flexibility in the apolipoprotein E amino-terminal domain structure determined from three new crystal forms: implications for lipid binding.
Protein Sci., 9, 2000
|
|
1B68
 
 | |
1AV1
 
 | |
