7V9Q
 
 | |
7V9P
 
 | |
7V9O
 
 | |
7V9N
 
 | | Crystal structure of the lanthipeptide zinc-metallopeptidase EryP from saccharopolyspora erythraea in closed state | | Descriptor: | ACETATE ION, Alanine aminopeptidase, CALCIUM ION, ... | | Authors: | Zhao, C, Zhao, N.L, Bao, R. | | Deposit date: | 2021-08-26 | | Release date: | 2022-05-11 | | Last modified: | 2022-07-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational remodeling enhances activity of lanthipeptide zinc-metallopeptidases.
Nat.Chem.Biol., 18, 2022
|
|
7SH0
 
 | | CRYSTAL STRUCTURE OF ENDOPLASMIC RETICULUM AMINOPEPTIDASE 2 (ERAP2) COMPLEX WITH A HIGHLY SELECTIVE AND POTENT SMALL MOLECULE | | Descriptor: | (2S)-N-hydroxy-3-(4-methoxyphenyl)-2-[4-({[5-(pyridin-2-yl)thiophene-2-sulfonyl]amino}methyl)-1H-1,2,3-triazol-1-yl]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Li, L, Bouvier, M. | | Deposit date: | 2021-10-07 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of the First Selective Nanomolar Inhibitors of ERAP2 by Kinetic Target-Guided Synthesis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7VPP
 
 | | Structures of a deltacoronavirus spike protein bound to porcine and human receptors indicate the risk of virus adaptation to humans | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ji, W, Xu, Y, Zhang, S. | | Deposit date: | 2021-10-17 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structures of a deltacoronavirus spike protein bound to porcine and human receptors.
Nat Commun, 13, 2022
|
|
7VPQ
 
 | | Structures of a deltacoronavirus spike protein bound to porcine and human receptors indicate the risk of virus adaptation to humans | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ji, W, Xu, Y, Zhang, S. | | Deposit date: | 2021-10-17 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of a deltacoronavirus spike protein bound to porcine and human receptors.
Nat Commun, 13, 2022
|
|
7U0L
 
 | | Crystal structure of the CCoV-HuPn-2018 RBD (domain B) in complex with canine APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7Z28
 
 | | High-resolution crystal structure of ERAP1 with bound bestatin analogue inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Papakyriakou, A, Stratikos, E, Vourloumis, D. | | Deposit date: | 2022-02-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Selective Nanomolar Inhibitors for Insulin-Regulated Aminopeptidase Based on alpha-Hydroxy-beta-amino Acid Derivatives of Bestatin.
J.Med.Chem., 65, 2022
|
|
7ZYF
 
 | | Insulin regulated aminopeptidase (IRAP) in complex with a nanomolar alpha hydroxy beta amino acid based inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Stratikos, E, Giastas, P, Papakyriakou, A. | | Deposit date: | 2022-05-24 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of Selective Nanomolar Inhibitors for Insulin-Regulated Aminopeptidase Based on alpha-Hydroxy-beta-amino Acid Derivatives of Bestatin.
J.Med.Chem., 65, 2022
|
|
8AVA
 
 | |
8AWH
 
 | |
8EX3
 
 | | Plasmodium falciparum M1 in complex with inhibitor 9aa | | Descriptor: | GLYCEROL, M1 family aminopeptidase, MAGNESIUM ION, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
8EWZ
 
 | | Plasmodium falciparum M1 in complex with inhibitor 9c | | Descriptor: | (2R)-2-(cyclopentylcarbamamido)-N-hydroxy-2-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)acetamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
8EYD
 
 | | Plasmodium falciparum M1 in complex with inhibitor 15ah | | Descriptor: | GLYCEROL, M1 family aminopeptidase, N-[(1R)-1-(4-bromophenyl)-2-(hydroxyamino)-2-oxoethyl]-N~2~-(4-fluorophenyl)glycinamide, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
8EYE
 
 | | Plasmodium falciparum M1 in complex with inhibitor 9aj | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
8EYF
 
 | | Plasmodium falciparum M1 in complex with inhibitor 15aa | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
8EZ2
 
 | | Plasmodium falciparum M1 in complex with inhibitor 15ag | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
8CGP
 
 | | Insulin regulated aminopeptidase (IRAP) in complex with an allosteric aryl sulfonamide inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Stratikos, E, Giastas, P. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Mechanisms of Allosteric Inhibition of Insulin-Regulated Aminopeptidase.
J.Mol.Biol., 436, 2024
|
|
8CGW
 
 | | Insulin-regulated aminopeptidase (IRAP) in complex with an allosteric benzopyran-based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Stratikos, E, Giastas, P. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Mechanisms of Allosteric Inhibition of Insulin-Regulated Aminopeptidase.
J.Mol.Biol., 436, 2024
|
|
8SLO
 
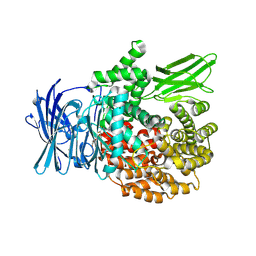 | | Plasmodium falciparum M1 aminopeptidase bound to selective inhibitor MIPS2673 | | Descriptor: | 2-hydroxy-N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]-2-methylpropanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | McGowan, S.M, Drinkwater, N. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Chemoproteomics validates selective targeting of Plasmodium M1 alanyl aminopeptidase as a cross-species strategy to treat malaria
Elife, 2024
|
|
8SW1
 
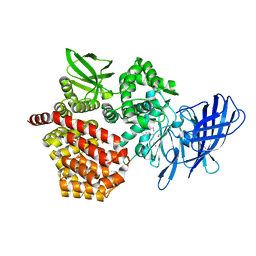 | |
8SW0
 
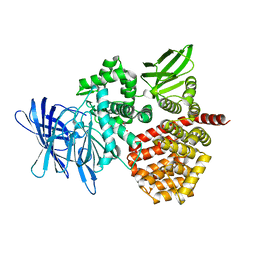 | | Puromycin sensitive aminopeptidase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Puromycin-sensitive aminopeptidase, ... | | Authors: | Rodgers, D.W, Sampath, S. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of puromycin-sensitive aminopeptidase and polyglutamine binding.
Plos One, 18, 2023
|
|
8T41
 
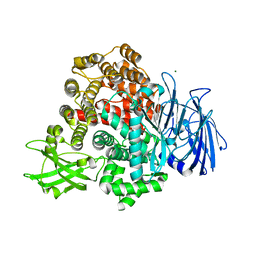 | |
8QOW
 
 | | LTA4 hydrolase in complex with compound 2(S) | | Descriptor: | (2~{S})-2-azanyl-3-[3-[4-[3-fluoranyl-5-(1~{H}-pyrazol-5-yl)pyridin-2-yl]oxyphenyl]pyrazol-1-yl]propan-1-ol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2023-09-29 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Amino Alcohols as Highly Potent, Selective, and Orally Efficacious Inhibitors of Leukotriene A4 Hydrolase.
J.Med.Chem., 66, 2023
|
|
