7MJQ
 
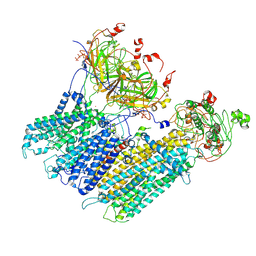 | | Vascular KATP channel: Kir6.1 SUR2B quatrefoil-like conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ATP-sensitive inward rectifier potassium channel 8, ... | | Authors: | Sung, M.W, Shyng, S.L. | | Deposit date: | 2021-04-20 | | Release date: | 2021-10-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Vascular K ATP channel structural dynamics reveal regulatory mechanism by Mg-nucleotides.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MJO
 
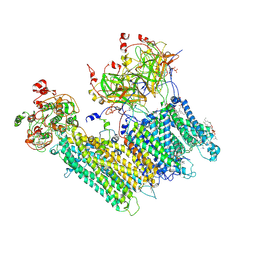 | | Vascular KATP channel: Kir6.1 SUR2B quatrefoil-like conformation 1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ... | | Authors: | Sung, M.W, Shyng, S.L. | | Deposit date: | 2021-04-20 | | Release date: | 2021-10-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Vascular K ATP channel structural dynamics reveal regulatory mechanism by Mg-nucleotides.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MIT
 
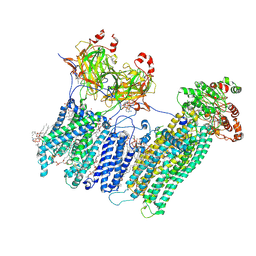 | | Vascular KATP channel: Kir6.1 SUR2B propeller-like conformation 1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ... | | Authors: | Sung, M.W, Shyng, S.L. | | Deposit date: | 2021-04-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Vascular K ATP channel structural dynamics reveal regulatory mechanism by Mg-nucleotides.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6XIT
 
 | | Cryo-EM structure of the G protein-gated inward rectifier K+ channel GIRK2 (Kir3.2) in complex with PIP2 | | Descriptor: | G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Niu, Y, Tao, X, MacKinnon, R. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM analysis of PIP 2 regulation in mammalian GIRK channels.
Elife, 9, 2020
|
|
6XIS
 
 | |
6XEV
 
 | | CryoEM structure of GIRK2-PIP2/CHS - G protein-gated inwardly rectifying potassium channel GIRK2 with modulators cholesteryl hemisuccinate and PIP2 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, ... | | Authors: | Mathiharan, Y.K, Glaaser, I.W, Skiniotis, G, Slesinger, P.A. | | Deposit date: | 2020-06-14 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into GIRK2 channel modulation by cholesterol and PIP2
Cell Rep, 36, 2021
|
|
6XEU
 
 | | CryoEM structure of GIRK2PIP2* - G protein-gated inwardly rectifying potassium channel GIRK2 with PIP2 | | Descriptor: | G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, SODIUM ION, ... | | Authors: | Mathiharan, Y.K, Glaaser, I.W, Skiniotis, G, Slesinger, P.A. | | Deposit date: | 2020-06-13 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into GIRK2 channel modulation by cholesterol and PIP2
Cell Rep, 36, 2021
|
|
6PZA
 
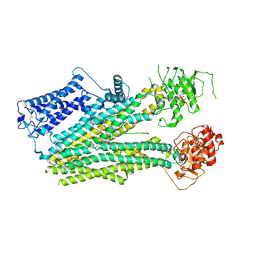 | | Cryo-EM structure of the pancreatic beta-cell SUR1 bound to ATP and glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Shyng, S.L, Yoshioka, C, Martin, G.M, Sung, M.W. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Mechanism of pharmacochaperoning in a mammalian K ATP channel revealed by cryo-EM.
Elife, 8, 2019
|
|
6PZ9
 
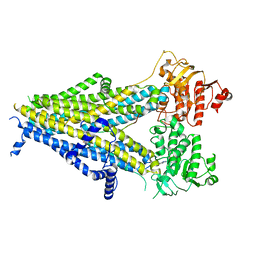 | | Cryo-EM structure of the pancreatic beta-cell SUR1 bound to ATP and repaglinide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ATP-sensitive inward rectifier potassium channel 11, ... | | Authors: | Shyng, S.L, Yoshioka, C, Martin, G.M, Sung, M.W. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Mechanism of pharmacochaperoning in a mammalian K ATP channel revealed by cryo-EM.
Elife, 8, 2019
|
|
6JB1
 
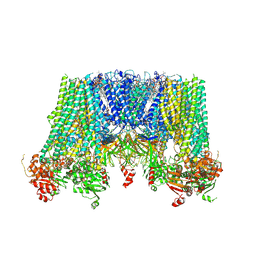 | | Structure of pancreatic ATP-sensitive potassium channel bound with repaglinide and ATPgammaS at 3.3A resolution | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ATP-binding cassette sub-family C member 8 isoform X2, ATP-sensitive inward rectifier potassium channel 11, ... | | Authors: | Chen, L, Ding, D, Wang, M, Wu, J.-X, Kang, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-22 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The Structural Basis for the Binding of Repaglinide to the Pancreatic KATPChannel.
Cell Rep, 27, 2019
|
|
6M84
 
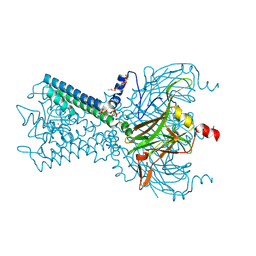 | | Crystal structure of cKir2.2 force open mutant in complex with PI(4,5)P2 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 12, DODECYL-BETA-D-MALTOSIDE, POTASSIUM ION, ... | | Authors: | Lee, S.-J, Ren, F, Yuan, P, Nichols, C.G. | | Deposit date: | 2018-08-21 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Atomistic basis of opening and conduction in mammalian inward rectifier potassium (Kir2.2) channels.
J.Gen.Physiol., 152, 2020
|
|
6M85
 
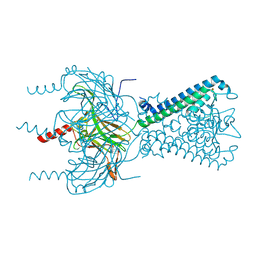 | |
6M86
 
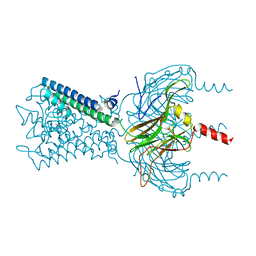 | |
6C3P
 
 | | Cryo-EM structure of human KATP bound to ATP and ADP in propeller form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Lee, K.P.K, Chen, J, MacKinnon, R. | | Deposit date: | 2018-01-10 | | Release date: | 2018-01-24 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Molecular structure of human KATP in complex with ATP and ADP.
Elife, 6, 2017
|
|
6C3O
 
 | | Cryo-EM structure of human KATP bound to ATP and ADP in quatrefoil form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Lee, K.P.K, Chen, J, MacKinnon, R. | | Deposit date: | 2018-01-10 | | Release date: | 2018-01-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular structure of human KATP in complex with ATP and ADP
Elife, 6, 2017
|
|
5YWC
 
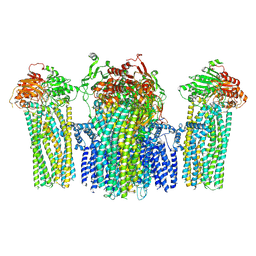 | |
5YW9
 
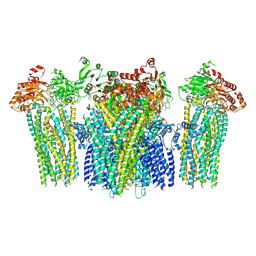 | |
5YWB
 
 | |
5YW8
 
 | |
5YWA
 
 | |
5YKF
 
 | |
5YKG
 
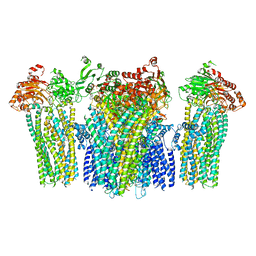 | |
5YKE
 
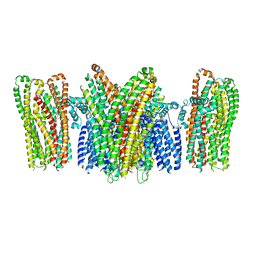 | |
6BAA
 
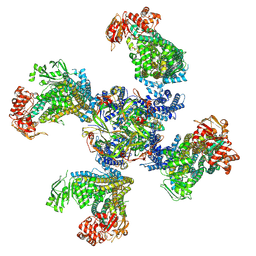 | | Cryo-EM structure of the pancreatic beta-cell KATP channel bound to ATP and glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Martin, G.M, Yoshioka, C, Shyng, S.L. | | Deposit date: | 2017-10-12 | | Release date: | 2017-11-01 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Anti-diabetic drug binding site in a mammalian KATPchannel revealed by Cryo-EM.
Elife, 6, 2017
|
|
5WUA
 
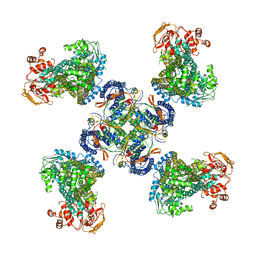 | | Structure of a Pancreatic ATP-sensitive Potassium Channel | | Descriptor: | ATP-sensitive inward rectifier potassium channel 11,superfolder GFP, SUR1 | | Authors: | Li, N, Wu, J.-X, Chen, L, Gao, N. | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-25 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure of a Pancreatic ATP-Sensitive Potassium Channel
Cell, 168, 2017
|
|
