4FSF
 
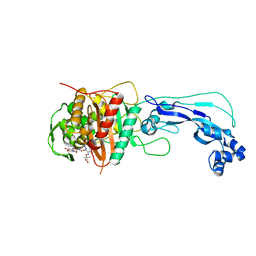 | | Crystal structure of Pseudomonas aeruginosa PBP3 complexed with compound 14 | | Descriptor: | (4R,5S,8Z)-8-(2-amino-1,3-thiazol-4-yl)-1-[3-(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)-1,2-oxazol-5-yl]-5-formyl-11,11-dimethyl-1,7-dioxo-4-(sulfoamino)-10-oxa-2,6,9-triazadodec-8-en-12-oic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel monobactams utilizing a siderophore uptake mechanism for the treatment of gram-negative infections.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3FWL
 
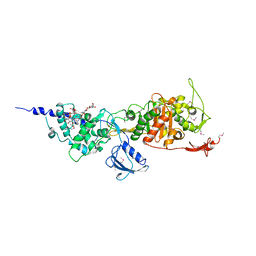 | | Crystal Structure of the Full-Length Transglycosylase PBP1b from Escherichia coli | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Sung, M.T, Lai, Y.T, Huang, C.Y, Chou, L.Y, Wong, C.H, Ma, C. | | Deposit date: | 2009-01-19 | | Release date: | 2009-06-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1QME
 
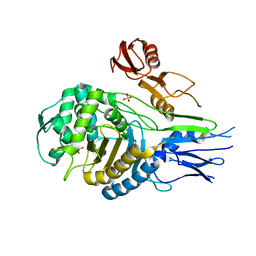 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2X, SULFATE ION | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Penicillin-Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance.
J.Mol.Biol., 299, 2000
|
|
1QMF
 
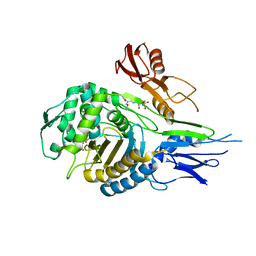 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) ACYL-ENZYME COMPLEX | | Descriptor: | 2-[CARBOXY-(2-FURAN-2-YL-2-METHOXYIMINO-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, CEFUROXIME (OCT-3-ENE FORM), PENICILLIN-BINDING PROTEIN 2X | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the Penicillin Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance
J.Mol.Biol., 299, 2000
|
|
1RP5
 
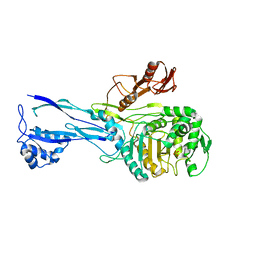 | | PBP2x from Streptococcus pneumoniae strain 5259 with reduced susceptibility to beta-lactam antibiotics | | Descriptor: | SULFATE ION, penicillin-binding protein 2x | | Authors: | Pernot, L, Chesnel, L, Legouellec, A, Croize, J, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to beta-lactam antibiotics.
J.Biol.Chem., 279, 2004
|
|
6G9S
 
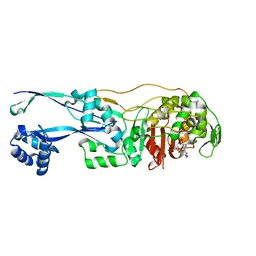 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | (3~{R},6~{S})-6-(aminomethyl)-4-(1,3-oxazol-5-yl)-3-(sulfooxyamino)-3,6-dihydro-2~{H}-pyridine-1-carboxylic acid, Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-11 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
6G9F
 
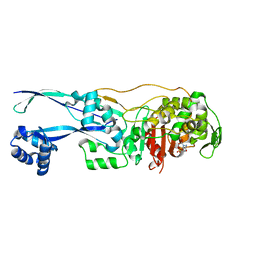 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
6G88
 
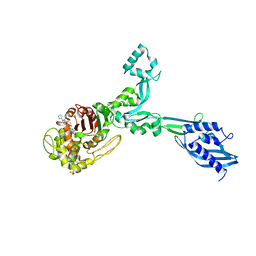 | | Crystal structure of Enterococcus Faecium D63r Penicillin-Binding protein 5 (PBP5fm) | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyrrolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Low affinity penicillin-binding protein 5 (PBP5), SULFATE ION | | Authors: | Sauvage, E, El Gachi, M, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of inactivation of Enterococcus faecium penicillin binding protein 5 by ceftobiprole.
To Be Published
|
|
6G9P
 
 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-11 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
1PMD
 
 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) | | Descriptor: | PEPTIDOGLYCAN SYNTHESIS MULTIFUNCTIONAL ENZYME | | Authors: | Pares, S, Mouz, N, Dideberg, O. | | Deposit date: | 1996-02-05 | | Release date: | 1997-02-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme.
Nat.Struct.Biol., 3, 1996
|
|
3OC2
 
 | | Crystal structure of penicillin-binding protein 3 from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, Penicillin-binding protein 3 | | Authors: | Sainsbury, S, Bird, L, Stuart, D.I, Owens, R.J, Ren, J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
J.Mol.Biol., 405, 2011
|
|
7OK9
 
 | |
7O4A
 
 | |
7O49
 
 | |
5UY7
 
 | |
8TJ3
 
 | |
8U55
 
 | |
7ZG8
 
 | |
7ZUI
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 5Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2R)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUH
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) Streptococcus pneumoniae R6 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Penicillin-binding protein 1b | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.467 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUJ
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 6Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2S)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUL
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with 8Az lactone - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2R)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-3-phenyl-propan-2-yl]hexanamide, CHLORIDE ION, Penicillin-binding protein 1b | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUK
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 7Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2S)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-3-phenyl-propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
1MWT
 
 | |
1MWR
 
 | |
