4BXZ
 
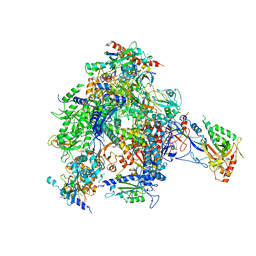 | | RNA Polymerase II-Bye1 complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-16 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6Q3M
 
 | |
7VVZ
 
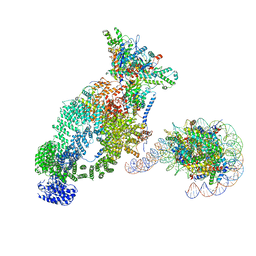 | | NuA4 bound to the nucleosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Qu, K, Chen, Z. | | Deposit date: | 2021-11-09 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structure of the NuA4 acetyltransferase complex bound to the nucleosome.
Nature, 610, 2022
|
|
7VVU
 
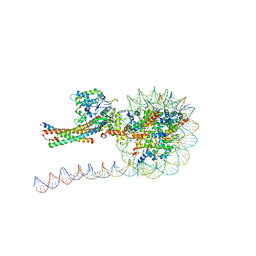 | | NuA4 HAT module bound to the nucleosome | | Descriptor: | CARBOXYMETHYL COENZYME *A, Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, ... | | Authors: | Chen, Z, Qu, K. | | Deposit date: | 2021-11-09 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the NuA4 acetyltransferase complex bound to the nucleosome.
Nature, 610, 2022
|
|
4COS
 
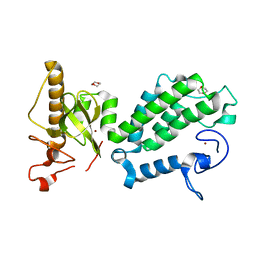 | | Crystal structure of the PHD-Bromo-PWWP cassette of human PRKCBP1 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, PROTEIN KINASE C-BINDING PROTEIN 1, ZINC ION | | Authors: | Krojer, T, Savitsky, P, Newman, J.A, Cooper, C.D.O, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Filippakopoulos, P. | | Deposit date: | 2014-01-30 | | Release date: | 2014-03-05 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Multivalent Histone and DNA Engagement by a PHD/BRD/PWWP Triple Reader Cassette Recruits ZMYND8 to K14ac-Rich Chromatin.
Cell Rep, 17, 2016
|
|
6UH5
 
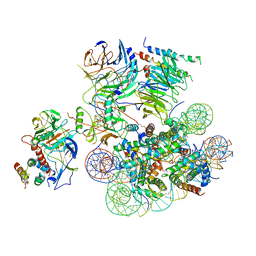 | | Structural basis of COMPASS eCM recognition of the H2Bub nucleosome | | Descriptor: | Bre2, DNA (146-MER), H3 N-terminus, ... | | Authors: | Hsu, P.L, Shi, H, Zheng, N. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS.
Mol.Cell, 76, 2019
|
|
6UGM
 
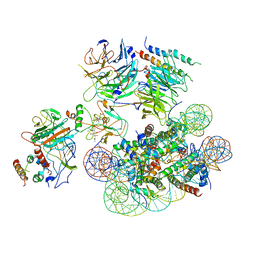 | | Structural basis of COMPASS eCM recognition of an unmodified nucleosome | | Descriptor: | Bre2, DNA (146-MER), H3 N-terminus, ... | | Authors: | Hsu, P.L, Shi, H, Zheng, N. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS.
Mol.Cell, 76, 2019
|
|
6VEN
 
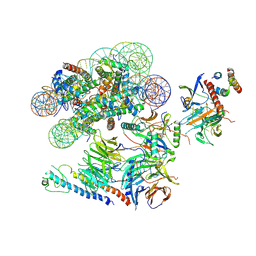 | |
5H1U
 
 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 2 | | Descriptor: | 2-amino-1,3-benzothiazole-6-carboxamide, DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ... | | Authors: | Liu, J. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
5H1T
 
 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 1 | | Descriptor: | DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ZINC ION, ... | | Authors: | Liu, J. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
5H1V
 
 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 6 | | Descriptor: | 2-Hydrazino-1,3-benzothiazole-6-carbohydrazide, DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ... | | Authors: | Liu, J. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
8GXQ
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in MH-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X, Xu, Y. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (5.04 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
8GXS
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in H-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
8I02
 
 | | Cryo-EM structure of the SIN3S complex from S. pombe | | Descriptor: | Chromatin modification-related protein eaf3, Cph1, Histone deacetylase clr6, ... | | Authors: | Wang, C, Guo, Z, Zhan, X. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Two assembly modes for SIN3 histone deacetylase complexes.
Cell Discov, 9, 2023
|
|
5B73
 
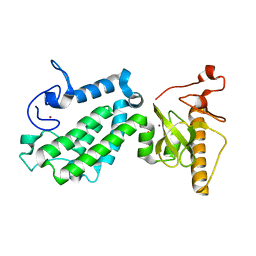 | | Crystal structure of human ZMYND8 PHD-Bromo-PWWP domain | | Descriptor: | Protein kinase C-binding protein 1, ZINC ION | | Authors: | Li, H, Li, Y, Zheng, X. | | Deposit date: | 2016-06-03 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ZMYND8 Reads the Dual Histone Mark H3K4me1-H3K14ac to Antagonize the Expression of Metastasis-Linked Genes
Mol.Cell, 63, 2016
|
|
6IET
 
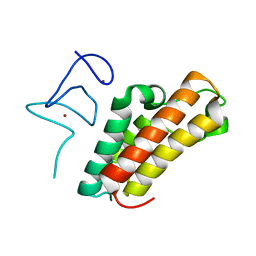 | | The crystal structure of TRIM66 PHD-Bromo domain | | Descriptor: | Tripartite motif-containing protein 66, ZINC ION | | Authors: | Chen, J. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | TRIM66 reads unmodified H3R2K4 and H3K56ac to respond to DNA damage in embryonic stem cells.
Nat Commun, 10, 2019
|
|
6IEU
 
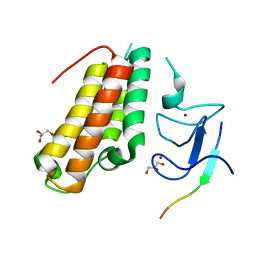 | |
3O36
 
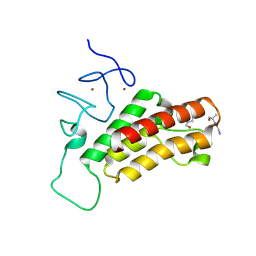 | |
3O35
 
 | |
3O33
 
 | |
3O34
 
 | |
3O37
 
 | |
6G8R
 
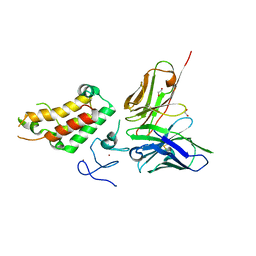 | | SP140 PHD-Bromodomain complex with scFv | | Descriptor: | 1,2-ETHANEDIOL, Nuclear body protein SP140, ZINC ION, ... | | Authors: | Fairhead, M, Graslund, S, Strain-Damerell, C, Picaud, S.S, Pike, A.C.W, Pinkas, D.M, Wigren, E, Preger, C, Persson Lotsholm, H, Ossipova, E, Filippakopoulos, P, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-09 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | SP140 PHD-Bromodomain complex with scFv
To Be Published
|
|
6LTH
 
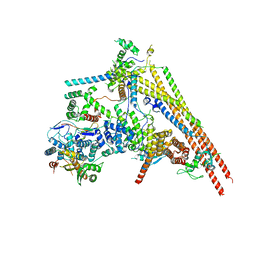 | | Structure of human BAF Base module | | Descriptor: | AT-rich interactive domain-containing protein 1A, SWI/SNF complex subunit SMARCC2, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1, ... | | Authors: | He, S, Wu, Z, Tian, Y, Yu, Z, Yu, J, Wang, X, Li, J, Liu, B, Xu, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-02-12 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of nucleosome-bound human BAF complex.
Science, 367, 2020
|
|
6GUU
 
 | |
