1MEI
 
 | |
4AVF
 
 | | Crystal structure of Pseudomonas aeruginosa inosine 5'-monophosphate dehydrogenase | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE | | Authors: | McMahon, S.A, Moynie, L, Liu, H, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-05-25 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1NF7
 
 | |
4DQW
 
 | | Crystal Structure Analysis of PA3770 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Labesse, G, Munier-Lehmann, H. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | MgATP Regulates Allostery and Fiber Formation in IMPDHs.
Structure, 21, 2013
|
|
1ME8
 
 | | Inosine Monophosphate Dehydrogenase (IMPDH) From Tritrichomonas Foetus with RVP bound | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, POTASSIUM ION, RIBAVIRIN MONOPHOSPHATE, ... | | Authors: | Prosise, G.L, Wu, J, Luecke, H. | | Deposit date: | 2002-08-08 | | Release date: | 2003-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Tritrichomonas foetus Inosine Monophosphate Dehydrogenase
in Complex with the Inhibitor Ribavirin Monophosphate Reveals a
Catalysis-dependent Ion-binding Site
J.Biol.Chem., 277, 2002
|
|
1NFB
 
 | |
4CFF
 
 | | Structure of full length human AMPK in complex with a small molecule activator, a thienopyridone derivative (A-769662) | | Descriptor: | 3-[4-(2-hydroxyphenyl)phenyl]-4-oxidanyl-6-oxidanylidene-7H-thieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Carmena, D, Bright, N.J, Haire, L.F, Underwood, E, Patel, B.R, Heath, R.B, Walker, P.A, Hallen, S, Giordanetto, F, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.924 Å) | | Cite: | Structural Basis of Ampk Regulation by Small Molecule Activators.
Nat.Commun., 4, 2013
|
|
4CFH
 
 | | Structure of an active form of mammalian AMPK | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2013-11-18 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
4FXS
 
 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae complexed with IMP and mycophenolic acid | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, MYCOPHENOLIC ACID, ... | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-07-03 | | Release date: | 2012-07-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae complexed with IMP and mycophenolic acid.
To be Published
|
|
4FEZ
 
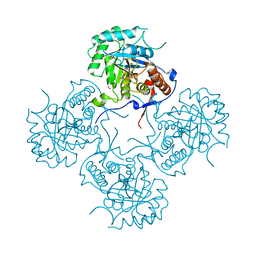 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant.
To be Published
|
|
7RES
 
 | | HUMAN IMPDH1 TREATED WITH ATP, IMP, AND NAD+, OCTAMER-CENTERED | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Isoform 5 of Inosine-5'-monophosphate dehydrogenase 1, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-13 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RGL
 
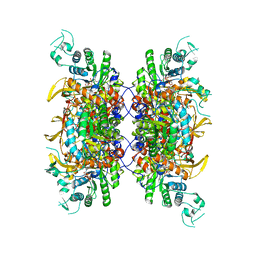 | | HUMAN RETINAL VARIANT IMPDH1(546) TREATED WITH ATP, IMP, NAD+, INTERFACE-CENTERED | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RFI
 
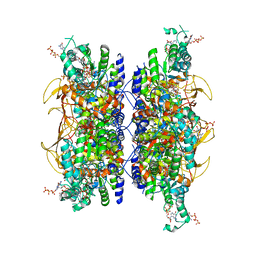 | | HUMAN RETINAL VARIANT IMPDH1(595) TREATED WITH GTP, ATP, IMP, NAD+, INTERFACE-CENTERED | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Isoform 5 of Inosine-5'-monophosphate dehydrogenase 1, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RFG
 
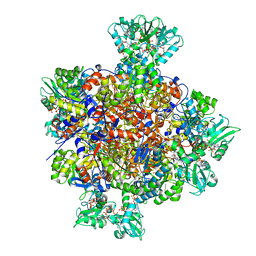 | | HUMAN IMPDH1 TREATED WITH GTP, IMP, AND NAD+ OCTAMER-CENTERED | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RFH
 
 | |
7RGQ
 
 | | HUMAN RETINAL VARIANT IMPDH1(546) TREATED WITH GTP, ATP, IMP, NAD+; INTERFACE-CENTERED | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RFF
 
 | |
7RFE
 
 | | HUMAN IMPDH1 TREATED WITH GTP, IMP, AND NAD+; INTERFACE-CENTERED | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RGM
 
 | | HUMAN RETINAL VARIANT IMPDH1(546) TREATED WITH ATP, IMP, NAD+, OCTAMER-CENTERED | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 1, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RER
 
 | | HUMAN IMPDH1 TREATED WITH ATP, IMP, AND NAD+ | | Descriptor: | INOSINIC ACID, Isoform 5 of Inosine-5'-monophosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-13 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RGI
 
 | | HUMAN RETINAL VARIANT IMPDH1(546) TREATED WITH GTP, ATP, IMP, NAD+; INTERFACE-CENTERED | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RGD
 
 | | HUMAN RETINAL VARIANT IMPDH1(595) TREATED WITH GTP, ATP, IMP, NAD+, OCTAMER-CENTERED | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6U8N
 
 | | Human IMPDH2 treated with ATP, IMP, and NAD+. Fully extended filament segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UDQ
 
 | | Human IMPDH2 treated with ATP, IMP, and 20 mM GTP. Fully compressed filament end reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-19 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6U8E
 
 | |
