3MT1
 
 | | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti | | Descriptor: | Putative carboxynorspermidine decarboxylase protein, SULFATE ION | | Authors: | Chang, C, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-30 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti
To be Published
|
|
3N29
 
 | | Crystal structure of carboxynorspermidine decarboxylase complexed with Norspermidine from Campylobacter jejuni | | Descriptor: | Carboxynorspermidine decarboxylase, GLYCEROL, N-(3-aminopropyl)propane-1,3-diamine, ... | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
7ODC
 
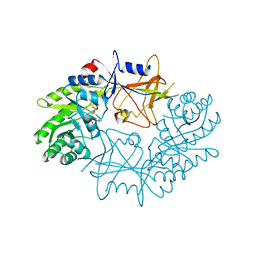 | | CRYSTAL STRUCTURE ORNITHINE DECARBOXYLASE FROM MOUSE, TRUNCATED 37 RESIDUES FROM THE C-TERMINUS, TO 1.6 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (ORNITHINE DECARBOXYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kern, A.D, Oliveira, M.A, Coffino, P, Hackert, M.L. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases.
Structure Fold.Des., 7, 1999
|
|
2J66
 
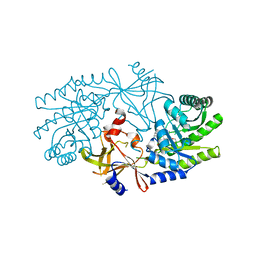 | | Structural characterisation of BtrK decarboxylase from butirosin biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, BTRK, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Popovic, B, Li, Y, Chirgadze, D.Y, Blundell, T.L, Spencer, J.B. | | Deposit date: | 2006-09-26 | | Release date: | 2006-09-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterisation of Btrk Decarboxylase from Bacillus Circulans Butirosin Biosynthesis
To be Published
|
|
4XG1
 
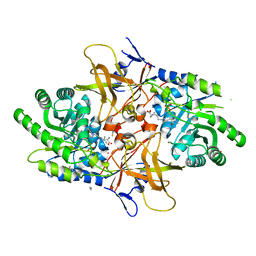 | | Psychromonas ingrahamii diaminopimelate decarboxylase with LLP | | Descriptor: | (2S)-2-amino-6-[[3-hydroxy-2-methyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]hexanoic acid, Diaminopimelate decarboxylase, POTASSIUM ION, ... | | Authors: | Peverelli, M.G, Wubben, J.M, Panjikar, S, Perugini, M.A. | | Deposit date: | 2014-12-30 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Expression to crystallization of diaminopimelate decarboxylase from the psychrophile Psychromonas ingrahamii
To Be Published
|
|
4AIB
 
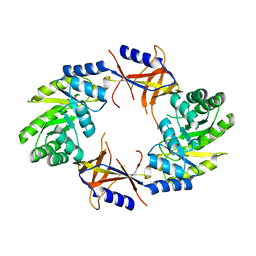 | |
1NJJ
 
 | |
3VAB
 
 | |
7JPJ
 
 | | Crystal Structure of the essential dimeric LYSA from Phaeodactylum tricornutum | | Descriptor: | D-LYSINE, Diaminopimelate decarboxylase, SULFATE ION | | Authors: | Fedorov, E, Belinski, V.A, Brunson, J.K, Almo, S.C, Dupont, C.L, Ghosh, A. | | Deposit date: | 2020-08-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The Phaeodactylum tricornutum diaminopimelate decarboxylase was acquired via horizontal gene transfer from bacteria and displays substrate promiscuity
Biorxiv, 2020
|
|
2QGH
 
 | | Crystal structure of diaminopimelate decarboxylase from Helicobacter pylori complexed with L-lysine | | Descriptor: | Diaminopimelate decarboxylase, GLYCEROL, LYSINE, ... | | Authors: | Hu, T, Wu, D, Jiang, H, Shen, X. | | Deposit date: | 2007-06-28 | | Release date: | 2008-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of diaminopimelate decarboxylase from Helicobacter pylori complexed with L-lysine
TO BE PUBLISHED
|
|
5X7M
 
 | |
7KH2
 
 | | Structure of N-citrylornithine decarboxylase bound with PLP | | Descriptor: | GLYCEROL, N-citrylornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Deng, X, Tomchick, D, Phillips, M, Michael, A. | | Deposit date: | 2020-10-19 | | Release date: | 2020-12-16 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Alternative pathways utilize or circumvent putrescine for biosynthesis of putrescine-containing rhizoferrin.
J.Biol.Chem., 296, 2020
|
|
1F3T
 
 | | CRYSTAL STRUCTURE OF TRYPANOSOMA BRUCEI ORNITHINE DECARBOXYLASE (ODC) COMPLEXED WITH PUTRESCINE, ODC'S REACTION PRODUCT. | | Descriptor: | 1,4-DIAMINOBUTANE, ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jackson, L.K, Brooks, H.B, Osterman, A.L, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2000-06-06 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Altering the reaction specificity of eukaryotic ornithine decarboxylase.
Biochemistry, 39, 2000
|
|
2TOD
 
 | | ORNITHINE DECARBOXYLASE FROM TRYPANOSOMA BRUCEI K69A MUTANT IN COMPLEX WITH ALPHA-DIFLUOROMETHYLORNITHINE | | Descriptor: | ALPHA-DIFLUOROMETHYLORNITHINE, PROTEIN (ORNITHINE DECARBOXYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Grishin, N.V, Osterman, A.L, Brooks, H.B, Phillips, M.A, Goldsmith, E.J. | | Deposit date: | 1999-05-18 | | Release date: | 1999-11-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with alpha-difluoromethylornithine.
Biochemistry, 38, 1999
|
|
7U6P
 
 | | Structure of an intellectual disability-associated ornithine decarboxylase variant G84R | | Descriptor: | Ornithine decarboxylase, PHOSPHATE ION | | Authors: | Zhou, X.E, Schultz, C.R, Powell, K.S, Henrickson, A, Lamp, J, Brunzelle, J.S, Demeler, B, Vega, I.E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Enzymatic Activity of an Intellectual Disability-Associated Ornithine Decarboxylase Variant, G84R.
Acs Omega, 7, 2022
|
|
7U6U
 
 | | Structure of an intellectual disability-associated ornithine decarboxylase variant G84R in complex with PLP | | Descriptor: | Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Schultz, C.R, Powell, K.S, Henrickson, A, Lamp, J, Brunzelle, J.S, Demeler, B, Vega, I.E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2022-03-06 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Enzymatic Activity of an Intellectual Disability-Associated Ornithine Decarboxylase Variant, G84R.
Acs Omega, 7, 2022
|
|
5X7N
 
 | |
1D7K
 
 | | CRYSTAL STRUCTURE OF HUMAN ORNITHINE DECARBOXYLASE AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN ORNITHINE DECARBOXYLASE | | Authors: | Almrud, J.J, Oliveira, M.A, Kern, A.D, Grishin, N.V, Phillips, M.A, Hackert, M.L. | | Deposit date: | 1999-10-18 | | Release date: | 2000-10-25 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human ornithine decarboxylase at 2.1 A resolution: structural insights to antizyme binding.
J.Mol.Biol., 295, 2000
|
|
2YXX
 
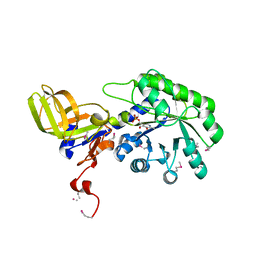 | |
4ZGY
 
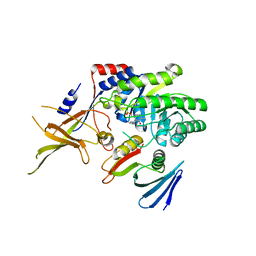 | |
7RU7
 
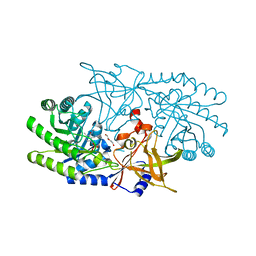 | | Crystal structure of BtrK, a decarboxylase involved in butirosin biosynthesis | | Descriptor: | DI(HYDROXYETHYL)ETHER, L-glutamyl-[BtrI acyl-carrier protein] decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Arenas, L.A.R, Paiva, F.C.R, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of BtrK, a decarboxylase involved in the (S)-4-amino-2-hydroxybutyrate (AHBA) formation during butirosin biosynthesis
J.Mol.Struct., 1267, 2022
|
|
7S3F
 
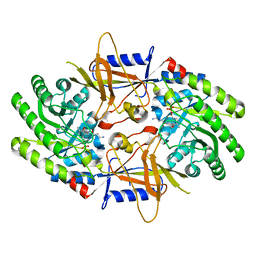 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with its inhibitor 1-amino-oxy-3-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
7S3G
 
 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with citrate at the catalytic center | | Descriptor: | CITRIC ACID, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
4ZGZ
 
 | |
3N2B
 
 | | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae. | | Descriptor: | CHLORIDE ION, Diaminopimelate decarboxylase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae.
To be Published
|
|
