5NIH
 
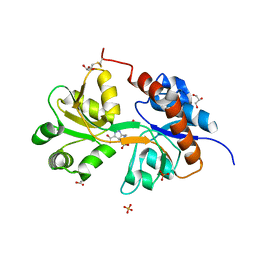 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with agonist LM-12b at 1.3 A resolution. | | 分子名称: | (3~{a}~{R},4~{S},6~{a}~{R})-1-methyl-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-c]pyrazole-3,4-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | 著者 | Laulumaa, S, Frydenvang, K.A, Kastrup, J.S. | | 登録日 | 2017-03-24 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
1SD3
 
 | |
6RUQ
 
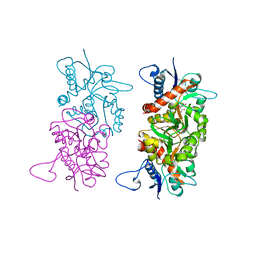
 | | Structure of GluA2cryst in complex the antagonist ZK200775 and the negative allosteric modulator GYKI53655 at 4.65 A resolution | | 分子名称: | (8R)-5-(4-aminophenyl)-N,8-dimethyl-8,9-dihydro-2H,7H-[1,3]dioxolo[4,5-h][2,3]benzodiazepine-7-carboxamide, Glutamate receptor 2, beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Krintel, C, Venskutonyte, R, Mirza, O.A, Gajhede, M, Kastrup, J.S. | | 登録日 | 2019-05-28 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (4.65 Å) | | 主引用文献 | Binding of a negative allosteric modulator and competitive antagonist can occur simultaneously at the ionotropic glutamate receptor GluA2.
Febs J., 288, 2021
|
|
5NEB
 
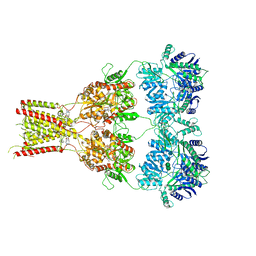
 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with LM-12b at 2.05 A resolution | | 分子名称: | (3~{a}~{R},4~{S},6~{a}~{R})-1-methyl-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-c]pyrazole-3,4-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | 著者 | Moellerud, S, Frydenvang, K, Laulumaa, S, Kastrup, J.S. | | 登録日 | 2017-03-10 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
5NG9
 
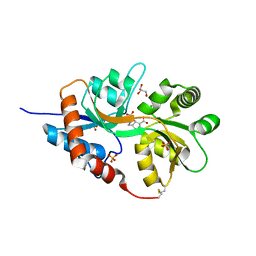
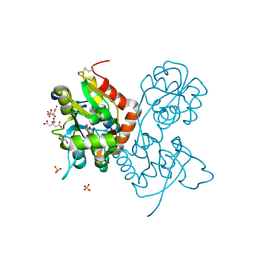 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with agonist CIP-AS at 1.15 A resolution. | | 分子名称: | (2~{S},3~{R},4~{R})-3-(carboxycarbonyl)-4-oxidanyl-pyrrolidine-2-carboxylic acid, (3~{a}~{S},4~{S},6~{a}~{R})-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-d][1,2]oxazole-3,4-dicarboxylic acid, CITRATE ANION, ... | | 著者 | Laulumaa, S, Frydenvang, K.A, Winther, S, Kastrup, J.S. | | 登録日 | 2017-03-17 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
6SBT
 
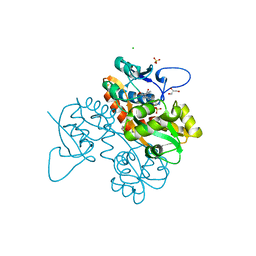 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with N-(7-(1H-imidazol-1-yl)-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl benzamide at 2.3 A resolution | | 分子名称: | CHLORIDE ION, GLYCEROL, Glutamate receptor ionotropic, ... | | 著者 | Moellerud, S, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2019-07-22 | | 公開日 | 2019-10-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | N-(7-(1H-Imidazol-1-yl)-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl)benzamide, a New Kainate Receptor Selective Antagonist and Analgesic: Synthesis, X-ray Crystallography, Structure-Affinity Relationships, and in Vitro and in Vivo Pharmacology.
Acs Chem Neurosci, 10, 2019
|
|
2CMO
 
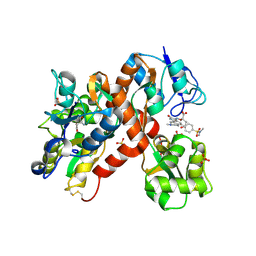 | | The structure of a mixed glur2 ligand-binding core dimer in complex with (s)-glutamate and the antagonist (s)-ns1209 | | 分子名称: | 2-({[(3E)-5-{4-[(DIMETHYLAMINO)(DIHYDROXY)-LAMBDA~4~-SULFANYL]PHENYL}-8-METHYL-2-OXO-6,7,8,9-TETRAHYDRO-1H-PYRROLO[3,2-H]ISOQUINOLIN-3(2H)-YLIDENE]AMINO}OXY)-4-HYDROXYBUTANOIC ACID, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | 著者 | Kasper, C, Pickering, D.S, Mirza, O, Olsen, L, Kristensen, A.S, Greenwood, J.R, Liljefors, T, Schousboe, A, Watjen, F, Gajhede, M, Sigurskjold, B.W, Kastrup, J.S. | | 登録日 | 2006-05-11 | | 公開日 | 2006-06-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | The Structure of a Mixed Glur2 Ligand-Binding Core Dimer in Complex with (S)-Glutamate and the Antagonist (S)-Ns1209.
J.Mol.Biol., 357, 2006
|
|
1S50
 
 | |
6R88
 
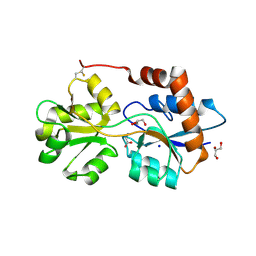 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with glycine | | 分子名称: | CHLORIDE ION, GLYCEROL, GLYCINE, ... | | 著者 | Alfieri, A, Pederzoli, R, Costa, A. | | 登録日 | 2019-04-01 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NF5
 
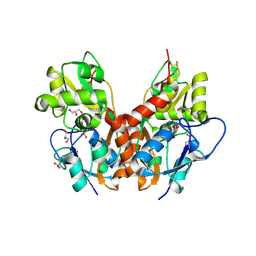 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with CIP-AS at 2.85 A resolution | | 分子名称: | (3~{a}~{S},4~{S},6~{a}~{R})-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-d][1,2]oxazole-3,4-dicarboxylic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Frydenvang, K, Venskutonyte, R, Thorsen, T.S, Kastrup, J.S. | | 登録日 | 2017-03-13 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
6QKC
 
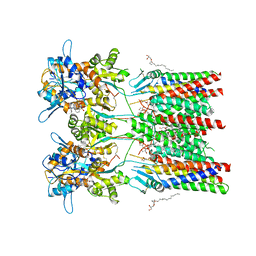 | | GluA1/2 In complex with auxiliary subunit gamma-8 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide, Glutamate receptor 1, ... | | 著者 | Herguedas, B, Garcia-Nafria, J, Greger, I.G. | | 登録日 | 2019-01-28 | | 公開日 | 2019-03-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP gamma 8.
Science, 364, 2019
|
|
1TXF
 
 | |
5O4F
 
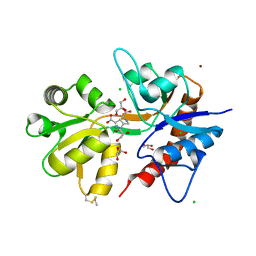 | | Structure of GluK3 ligand-binding domain (S1S2) in complex with the agonist LM-12b at 2.10 A resolution | | 分子名称: | (3~{a}~{R},4~{S},6~{a}~{R})-1-methyl-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-c]pyrazole-3,4-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | 著者 | Moellerud, S, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2017-05-29 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
2AL5
 
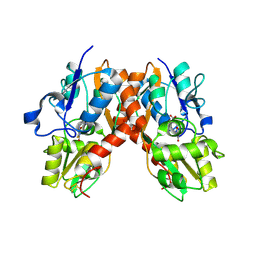 | | Crystal structure of the GluR2 ligand binding core (S1S2J) in complex with fluoro-willardiine and aniracetam | | 分子名称: | 1-(4-METHOXYBENZOYL)-2-PYRROLIDINONE, 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2 | | 著者 | Jin, R, Clark, S, Weeks, A.M, Dudman, J.T, Gouaux, E, Partin, K.M. | | 登録日 | 2005-08-04 | | 公開日 | 2005-10-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Mechanism of positive allosteric modulators acting on AMPA receptors.
J.Neurosci., 25, 2005
|
|
2ANJ
 
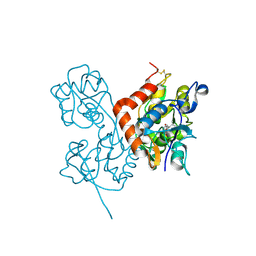 | | Crystal Structure of the Glur2 Ligand Binding Core (S1S2J-Y450W) Mutant in Complex With the Partial Agonist Kainic Acid at 2.1 A Resolution | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | 著者 | Holm, M.M, Naur, P, Vestergaard, B, Geballe, M.T, Gajhede, M, Kastrup, J.S, Traynelis, S.F, Egebjerg, J. | | 登録日 | 2005-08-11 | | 公開日 | 2005-08-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A Binding Site Tyrosine Shapes Desensitization Kinetics and Agonist Potency at GluR2: a mutagenic, kinetic, and crystallographic study
J.Biol.Chem., 280, 2005
|
|
5NS9
 
 | |
6R8A
 
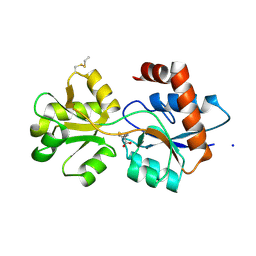 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-methionine | | 分子名称: | Glutamate receptor 3.3,Glutamate receptor 3.3, METHIONINE, SODIUM ION, ... | | 著者 | Alfieri, A, Pederzoli, R, Costa, A. | | 登録日 | 2019-04-01 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1TT1
 
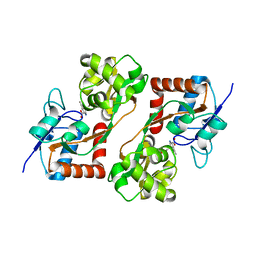 | |
2A5T
 
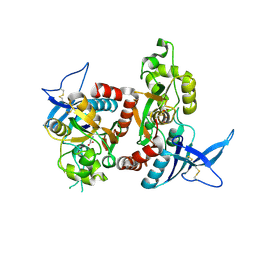 | | Crystal Structure Of The NR1/NR2A ligand-binding cores complex | | 分子名称: | GLUTAMIC ACID, GLYCINE, N-methyl-D-aspartate receptor NMDAR1-4a subunit, ... | | 著者 | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | 登録日 | 2005-06-30 | | 公開日 | 2005-11-15 | | 最終更新日 | 2017-07-26 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
5O9A
 
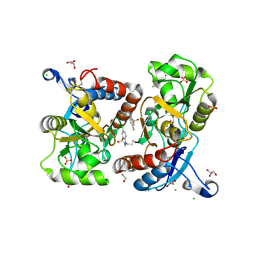 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L504Y-N775S) in complex with glutamate and BPAM121 at 1.78 A resolution | | 分子名称: | 1,2-ETHANEDIOL, 7-chloro-4-(2-fluoroethyl)-2,3-dihydro-1,2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, ... | | 著者 | Laulumaa, S, Rovinskaja, K, Frydenvang, K.A, Kastrup, J.S. | | 登録日 | 2017-06-16 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | 7-Phenoxy-Substituted 3,4-Dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides as Positive Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors with Nanomolar Potency.
J. Med. Chem., 61, 2018
|
|
2A5S
 
 | | Crystal Structure Of The NR2A Ligand Binding Core In Complex With Glutamate | | 分子名称: | GLUTAMIC ACID, N-methyl-D-aspartate receptor NMDAR2A subunit | | 著者 | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | 登録日 | 2005-06-30 | | 公開日 | 2005-11-15 | | 最終更新日 | 2017-07-26 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
5OEW
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with glutamate and positive allosteric modulator BPAM538 | | 分子名称: | 4-cyclopropyl-7-(3-methoxyphenoxy)-2,3-dihydro-1$l^{6},2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, GLUTAMIC ACID, ... | | 著者 | Larsen, A.P, Frydenvang, K.A, Kastrup, J.S. | | 登録日 | 2017-07-10 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 7-Phenoxy-Substituted 3,4-Dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides as Positive Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors with Nanomolar Potency.
J. Med. Chem., 61, 2018
|
|
1SYH
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 1.85 A RESOLUTION. | | 分子名称: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | 著者 | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | 登録日 | 2004-04-01 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
2AL4
 
 | | CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH quisqualate and CX614. | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2,3,6A,7,8,9-HEXAHYDRO-11H-[1,4]DIOXINO[2,3-G]PYRROLO[2,1-B][1,3]BENZOXAZIN-11-ONE, Glutamate receptor 2, ... | | 著者 | Jin, R, Clark, S, Weeks, A.M, Dudman, J.T, Gouaux, E, Partin, K.M. | | 登録日 | 2005-08-04 | | 公開日 | 2005-10-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mechanism of positive allosteric modulators acting on AMPA receptors.
J.Neurosci., 25, 2005
|
|
1S9T
 
 | |
