7T4R
 
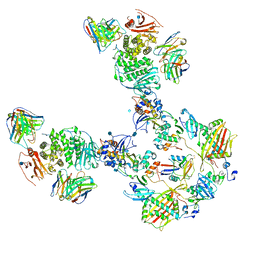 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with THBD and neutralizing fabs MSL-109 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7R8U
 
 | |
7QSR
 
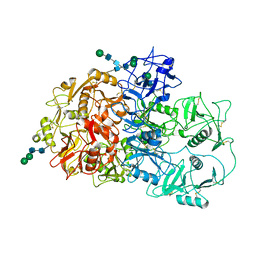 | | CryoEM structure of the Ectodomain of Human PLA2R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Secretory phospholipase A2 receptor, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Briggs, D.C, Lockhart-Cairns, M.P, Baldock, C. | | Deposit date: | 2022-01-14 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of PLA2R reveals presentation of the dominant membranous nephropathy epitope and an immunogenic patch.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7QAJ
 
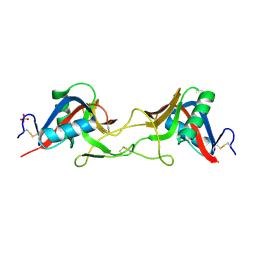 | | ZK002 with Anti-angiogenic and Anti-inflamamtory Properties | | Descriptor: | SULFATE ION, Snaclec clone 2100755 alpha, Snaclec clone 2100755 beta | | Authors: | Wong, W.Y, Chan, B.D, Muk Lan Lee, M, Dai, X, Tsim, K.W.K, Hsiao, W.L.W, Li, M, Li, X.Y, Tai, W.C.S. | | Deposit date: | 2021-11-17 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isolation and characterization of ZK002, a novel dual function snake venom protein from Deinagkistrodon acutus with anti-angiogenic and anti-inflammatory properties.
Front Pharmacol, 14, 2023
|
|
7ODU
 
 | | Natural killer cell receptor NKR-P1B from Rattus norvegicus in complex with its cognate ligand Clr-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D11, ... | | Authors: | Skalova, T, Blaha, J, Kalouskova, B, Skorepa, O, Vanek, O, Dohnalek, J. | | Deposit date: | 2021-04-30 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Natural killer cell receptor NKR-P1B from Rattus norvegicus in complex with its cognate ligand Clr-11
To Be Published
|
|
7NL7
 
 | | Crystal Structure of DC-SIGN in complex with a triazole-based glycomimetic ligand | | Descriptor: | 3-Aminopropyl 2-deoxy-2-(4-phenyl-1,2,3-triazol-1-yl)-alpha-D-mannopyranoside, CALCIUM ION, DC-SIGN, ... | | Authors: | Jakob, R.P, Cramer, J, Lakkaichi, A, Aliu, B, Cattaneo, I, Klein, S, Jiang, X, Rabbani, S, Schwardt, O, Ernst, B, Maier, T. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sweet Drugs for Bad Bugs: A Glycomimetic Strategy against the DC-SIGN-Mediated Dissemination of SARS-CoV-2.
J.Am.Chem.Soc., 143, 2021
|
|
7NL6
 
 | | Crystal Structure of DC-SIGN in complex with a triazole-based glycomimetic ligand | | Descriptor: | CALCIUM ION, DC-SIGN, CRD domain, ... | | Authors: | Jakob, R.P, Cramer, J, Lakkaichi, A, Aliu, B, Cattaneo, I, Klein, S, Jiang, X, Rabbani, S, Schwardt, O, Ernst, B, Maier, T. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sweet Drugs for Bad Bugs: A Glycomimetic Strategy against the DC-SIGN-Mediated Dissemination of SARS-CoV-2.
J.Am.Chem.Soc., 143, 2021
|
|
7L68
 
 | |
7L67
 
 | |
7L66
 
 | |
7L65
 
 | |
7L64
 
 | |
7L63
 
 | |
7L62
 
 | |
7L61
 
 | |
7JUH
 
 | |
7JUG
 
 | |
7JUF
 
 | |
7JUE
 
 | |
7JUD
 
 | |
7JUC
 
 | |
7JUB
 
 | |
7JPU
 
 | |
7JPT
 
 | | Structure of an endocytic receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 75, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gully, B.S, Rossjohn, J, Berry, R. | | Deposit date: | 2020-08-09 | | Release date: | 2020-12-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The cryo-EM structure of the endocytic receptor DEC-205.
J.Biol.Chem., 296, 2020
|
|
7FI9
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | GLYCEROL, MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
