2F2W
 
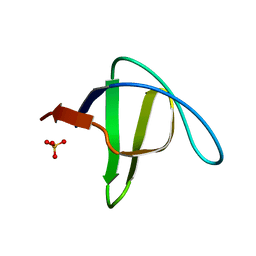 | | alpha-spectrin SH3 domain R21A mutant | | Descriptor: | SULFATE ION, Spectrin alpha chain, brain | | Authors: | Camara-Artigas, A, Conejero-Lara, F, Casares, S, Lopez-Mayorga, O, Vega, C. | | Deposit date: | 2005-11-18 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cooperative propagation of local stability changes from low-stability and high-stability regions in a SH3 domain
Proteins, 67, 2007
|
|
3M0T
 
 | |
7PW0
 
 | |
2W0Z
 
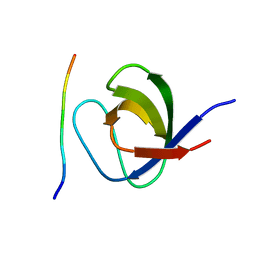 | | Grb2 SH3C (3) | | Descriptor: | GRB2-ASSOCIATED-BINDING PROTEIN 2, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2 | | Authors: | Harkiolaki, M, Tsirka, T, Feller, S.M. | | Deposit date: | 2008-10-13 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Distinct Binding Modes of Two Epitopes in Gab2 that Interact with the Sh3C Domain of Grb2.
Structure, 17, 2009
|
|
1UJ0
 
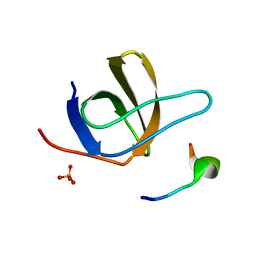 | | Crystal Structure of STAM2 SH3 domain in complex with a UBPY-derived peptide | | Descriptor: | PHOSPHATE ION, deubiquitinating enzyme UBPY, signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 | | Authors: | Kaneko, T, Kumasaka, T, Ganbe, T, Sato, T, Miyazawa, K, Kitamura, N, Tanaka, N. | | Deposit date: | 2003-07-24 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into modest binding of a non-PXXP ligand to the signal transducing adaptor molecule-2 Src homology 3 domain.
J.Biol.Chem., 278, 2003
|
|
3C0C
 
 | |
3THK
 
 | | Structure of SH3 chimera with a type II ligand linked to the chain C-terminal | | Descriptor: | BETA-MERCAPTOETHANOL, Proline-rich peptide, SULFATE ION, ... | | Authors: | Gabdulkhakov, A.G, Gushchina, L.V, Nikulin, A.D, Nikonov, S.V, Filimonov, V.V. | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-Resolution Crystal Structure of Spectrin SH3 Domain Fused with a Proline-Rich Peptide.
J.Biomol.Struct.Dyn., 29, 2011
|
|
7A2U
 
 | |
4J9H
 
 | |
1YN8
 
 | |
4J9B
 
 | |
5XGG
 
 | |
5H0H
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-N,N,4-trimethylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-~{N},~{N},4-trimethyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
4RUG
 
 | |
3M0Q
 
 | |
2O88
 
 | |
6ATV
 
 | | The molecular mechanisms by which NS1 of the 1918 Spanish influenza A virus hijack host protein-protein interactions | | Descriptor: | Adapter molecule crk, proline-rich motif in IAV-NS1 | | Authors: | Shen, Q, Zeng, D, Zhao, B, Li, P, Cho, J.H. | | Deposit date: | 2017-08-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Molecular Mechanisms of Tight Binding through Fuzzy Interactions.
Biophys. J., 114, 2018
|
|
3H0H
 
 | |
1OEB
 
 | | Mona/Gads SH3C domain | | Descriptor: | CADMIUM ION, GRB2-RELATED ADAPTOR PROTEIN 2, LYMPHOCYTE CYTOSOLIC PROTEIN 2 | | Authors: | Harkiolaki, M, Lewitzky, M, Gilbert, R.J.C, Jones, E.Y, Bourette, R.P, Mouchiroud, G, Sondermann, H, Moarefi, I, Feller, S.M. | | Deposit date: | 2003-03-24 | | Release date: | 2003-04-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for SH3 Domain-Mediated High-Affinity Binding between Mona/Gads and Slp-76
Embo J., 22, 2003
|
|
5L23
 
 | |
1TUD
 
 | |
1PWT
 
 | | THERMODYNAMIC ANALYSIS OF ALPHA-SPECTRIN SH3 AND TWO OF ITS CIRCULAR PERMUTANTS WITH DIFFERENT LOOP LENGTHS: DISCERNING THE REASONS FOR RAPID FOLDING IN PROTEINS | | Descriptor: | ALPHA SPECTRIN | | Authors: | Martinez, J.C, Viguera, A.R, Berisio, R, Wilmanns, M, Mateo, P.L, Filmonov, V.V, Serrano, L. | | Deposit date: | 1998-10-06 | | Release date: | 1999-05-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Thermodynamic analysis of alpha-spectrin SH3 and two of its circular permutants with different loop lengths: discerning the reasons for rapid folding in proteins.
Biochemistry, 38, 1999
|
|
5XG9
 
 | | Crystal Structure of PEG-bound SH3 domain of Myosin IB from Entamoeba histolytica | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, PENTAETHYLENE GLYCOL, ... | | Authors: | Gautam, G, Gourinath, S. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the PEG-bound SH3 domain of myosin IB from Entamoeba histolytica reveals its mode of ligand recognition
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5JMQ
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with CP3 | | Descriptor: | 1,2-ETHANEDIOL, 9-[(5E)-7-carbamimidamido-5,6,7-trideoxy-beta-D-ribo-hept-5-enofuranosyl]-9H-purin-6-amine, CALCIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
Febs J., 284, 2017
|
|
5OB2
 
 | |
