7PSN
 
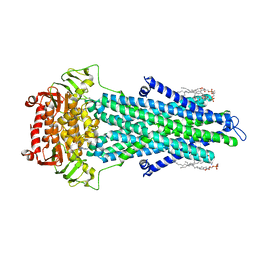 | |
7OTI
 
 | | Structure of ABCB1/P-glycoprotein in apo state | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ford, R.C, Barbieri, A, Thonghin, N, Shafi, T, Prince, S.M, Collins, R.F. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of ABCB1/P-Glycoprotein in the Presence of the CFTR Potentiator Ivacaftor.
Membranes (Basel), 11, 2021
|
|
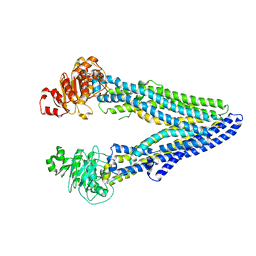
7OTG
 
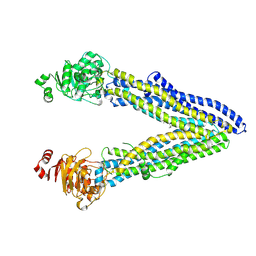 | | Structure of ABCB1/P-glycoprotein in the presence of the CFTR potentiator ivacaftor | | Descriptor: | Multidrug resistance protein 1A, N-(2,4-di-tert-butyl-5-hydroxyphenyl)-4-oxo-1,4-dihydroquinoline-3-carboxamide | | Authors: | Ford, R.C, Barbieri, A, Thonghin, N, Shafi, T, Prince, S.M, Collins, R.F. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structure of ABCB1/P-Glycoprotein in the Presence of the CFTR Potentiator Ivacaftor.
Membranes (Basel), 11, 2021
|
|
7S5X
 
 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S5V
 
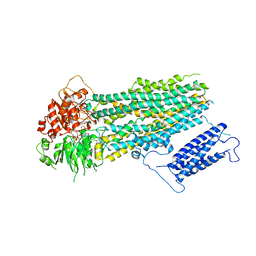 | | Human KATP channel in open conformation, focused on SUR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S5Y
 
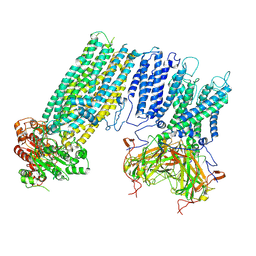 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S61
 
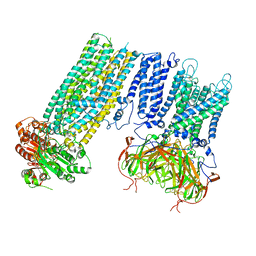 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S5Z
 
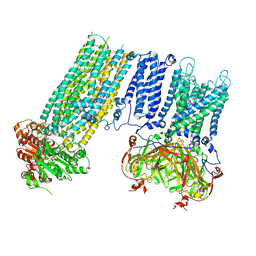 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S60
 
 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
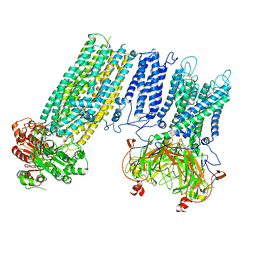
7LB8
 
 | | Structure of a ferrichrome importer FhuCDB from E. coli | | Descriptor: | Iron(3+)-hydroxamate import ATP-binding protein FhuC, Iron(3+)-hydroxamate import system permease protein FhuB, Iron(3+)-hydroxamate-binding protein FhuD | | Authors: | Hu, W, Zheng, H. | | Deposit date: | 2021-01-07 | | Release date: | 2021-11-24 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM reveals unique structural features of the FhuCDB Escherichia coli ferrichrome importer.
Commun Biol, 4, 2021
|
|
7P06
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in outward-facing conformation with ADP-orthovanadate/ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P05
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP and rhodamine 6G | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P03
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation without nucleotides | | Descriptor: | Pleiotropic ABC efflux transporter of multiple drugs | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P04
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7SHN
 
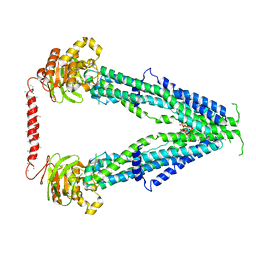 | | Cryo-EM structure of oleoyl-CoA-bound human peroxisomal fatty acid transporter ABCD1 | | Descriptor: | ATP-binding cassette sub-family D member 1, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | Authors: | Wang, R, Li, X. | | Deposit date: | 2021-10-09 | | Release date: | 2021-11-03 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of acyl-CoA transport across the peroxisomal membrane by human ABCD1.
Cell Res., 32, 2022
|
|
7SHM
 
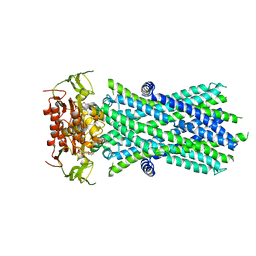 | |
7MJP
 
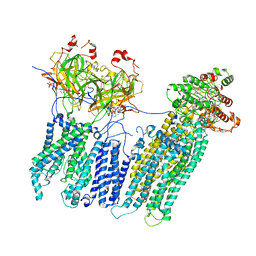 | | Vascular KATP channel: Kir6.1 SUR2B propeller-like conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Sung, M.W, Shyng, S.L. | | Deposit date: | 2021-04-20 | | Release date: | 2021-10-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Vascular K ATP channel structural dynamics reveal regulatory mechanism by Mg-nucleotides.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MJO
 
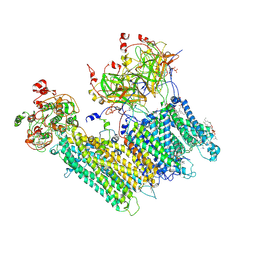 | | Vascular KATP channel: Kir6.1 SUR2B quatrefoil-like conformation 1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ... | | Authors: | Sung, M.W, Shyng, S.L. | | Deposit date: | 2021-04-20 | | Release date: | 2021-10-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Vascular K ATP channel structural dynamics reveal regulatory mechanism by Mg-nucleotides.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MJQ
 
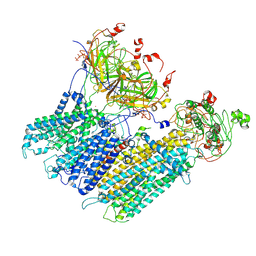 | | Vascular KATP channel: Kir6.1 SUR2B quatrefoil-like conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ATP-sensitive inward rectifier potassium channel 8, ... | | Authors: | Sung, M.W, Shyng, S.L. | | Deposit date: | 2021-04-20 | | Release date: | 2021-10-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Vascular K ATP channel structural dynamics reveal regulatory mechanism by Mg-nucleotides.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MIT
 
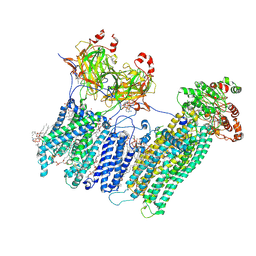 | | Vascular KATP channel: Kir6.1 SUR2B propeller-like conformation 1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ... | | Authors: | Sung, M.W, Shyng, S.L. | | Deposit date: | 2021-04-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Vascular K ATP channel structural dynamics reveal regulatory mechanism by Mg-nucleotides.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MEW
 
 | | E. coli MsbA in complex with G247 | | Descriptor: | (2E)-3-{1-cyclopropyl-7-[(1S)-1-(3,6-dichloro-2-fluorophenyl)ethoxy]naphthalen-2-yl}prop-2-enoic acid, ATP-dependent lipid A-core flippase | | Authors: | Thelot, F, Liao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-10-06 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Distinct allosteric mechanisms of first-generation MsbA inhibitors.
Science, 374, 2021
|
|
7MET
 
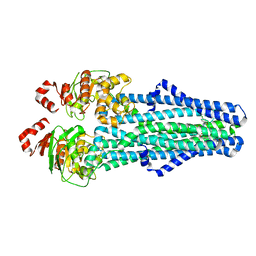 | | A. baumannii MsbA in complex with TBT1 decoupler | | Descriptor: | 2-(4-chlorobenzamido)-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Thelot, F, Liao, M. | | Deposit date: | 2021-04-07 | | Release date: | 2021-10-06 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Distinct allosteric mechanisms of first-generation MsbA inhibitors.
Science, 374, 2021
|
|
7RIT
 
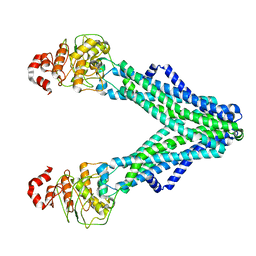 | | Drug-free A. baumannii MsbA | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Thelot, F, Liao, M. | | Deposit date: | 2021-07-20 | | Release date: | 2021-10-06 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Distinct allosteric mechanisms of first-generation MsbA inhibitors.
Science, 374, 2021
|
|
7CH9
 
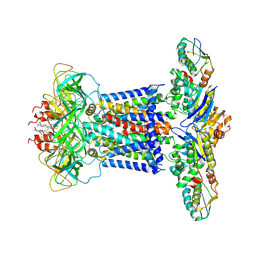 | | Cryo-EM structure of P.aeruginosa MlaFEBD | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, MlaD domain-containing protein, Probable ATP-binding component of ABC transporter, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7CH8
 
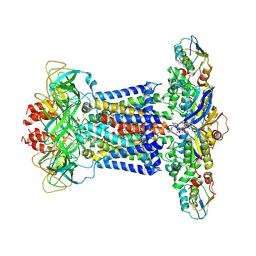 | | Cryo-EM structure of P.aeruginosa MlaFEBD with ADP-V | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
