8TO2
 
 | | Bottom cylinder of high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
8TO5
 
 | | Central rod disk in C1 symmetry of high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (1.87 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
8Q1R
 
 | | mouse Keap1 in complex with stapled peptide | | Descriptor: | Kelch-like ECH-associated protein 1, SODIUM ION, SULFATE ION, ... | | Authors: | Kack, H, Wissler, L. | | Deposit date: | 2023-08-01 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A cell-active cyclic peptide targeting the Nrf2/Keap1 protein-protein interaction.
Chem Sci, 14, 2023
|
|
8Q1S
 
 | | Pathogenic mutations of human phosphorylation sites affect protein-protein interactions | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein epsilon, BROMIDE ION, ... | | Authors: | Roske, Y, Daumke, O, Rrustemi, T, Selbach, M. | | Deposit date: | 2023-08-01 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Pathogenic mutations of human phosphorylation sites affect protein-protein interactions.
Nat Commun, 15, 2024
|
|
8Q1W
 
 | |
8TNH
 
 | |
8TNI
 
 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing bi-specific antibody CAP256L-R27 targeting the CD4-binding site and the V2-apex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 BG505 DS-SOSIP gp120, ... | | Authors: | Zhou, T, Morano, N.C, Roark, R.S, Kwong, P.D, Xu, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing bi-specific antibody CAP256L-R27 targeting the CD4-binding site and the V2-apex
To Be Published
|
|
8TNG
 
 | |
8Q1B
 
 | | III2-IV1 respiratory supercomplex from S. pombe | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Moe, A, Brzezinski, P. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of the S. pombe III-IV-cyt c supercomplex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8TMT
 
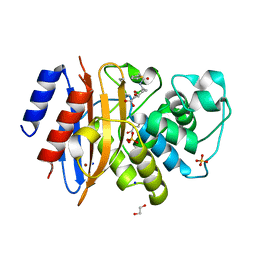 | | Crystal structure of KPC-44 carbapenemase in complex with vaborbactam | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, LITHIUM ION, ... | | Authors: | Sun, Z, Palzkill, T, Hu, L, Neetu, N, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-30 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8Q0S
 
 | | X-ray structure of the single chain monellin derivative MNEI | | Descriptor: | ACETATE ION, GLYCEROL, Monellin chain B,Monellin chain A, ... | | Authors: | Ferraro, G, Merlino, A, Lucignano, R, Picone, D. | | Deposit date: | 2023-07-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural insights and aggregation propensity of a super-stable monellin mutant: A new potential building block for protein-based nanostructured materials.
Int.J.Biol.Macromol., 254, 2024
|
|
8PZO
 
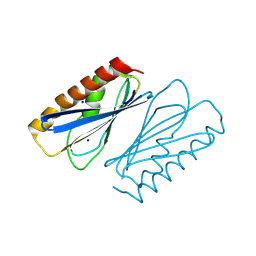 | | LpdD | | Descriptor: | Protein LpdD, SODIUM ION | | Authors: | Gahloth, D, Leys, D. | | Deposit date: | 2023-07-27 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of LpdD from Lactobacillus plantarum.
To Be Published
|
|
8PZY
 
 | | Intracellular leucine aminopeptidase of Pseudomonas aeruginosa PA14 - hexameric assembly with manganese bound | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-[3-(2-hydroxyethoxy)propoxy]propoxy]ethanol, AMMONIUM ION, ... | | Authors: | Simpson, M.C, Czekster, C.M, Harding, C.J. | | Deposit date: | 2023-07-27 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Unveiling the Catalytic Mechanism of a Processive Metalloaminopeptidase.
Biochemistry, 62, 2023
|
|
8Q00
 
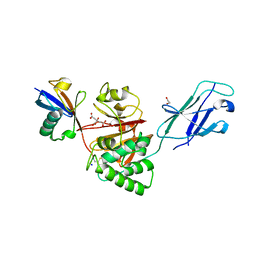 | | TssM-Ub-PA complex - A USP-like DUB from B. pseudomallei (193-430) reacted with Ub-PA | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Polyubiquitin-B, ... | | Authors: | Uthoff, M, Hermanns, T, Hofmann, K, Baumann, U. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structural basis for deubiquitination by the fingerless USP-type effector TssM.
Life Sci Alliance, 7, 2024
|
|
8K7X
 
 | | Crystal structure of GH146 beta-L-arabinofuranosidase Bll3HypBA1 (amino acids 380-1223) in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Pan, L, Maruyama, S, Miyake, M, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-07-27 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Bifidobacterial GH146 beta-L-arabinofuranosidase for the removal of beta 1,3-L-arabinofuranosides on plant glycans.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
8PYS
 
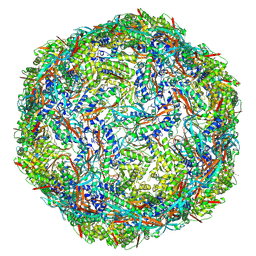 | |
8PZ0
 
 | | Intracellular leucine aminopeptidase of Pseudomonas aeruginosa PA14. | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Simpson, M.C, Czekster, C.M, Harding, C.J. | | Deposit date: | 2023-07-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unveiling the Catalytic Mechanism of a Processive Metalloaminopeptidase.
Biochemistry, 62, 2023
|
|
8PYW
 
 | | Crystal structure of the human Nucleoside-diphosphate kinase B domain bound to compound diphosphate form of AT-9052-Sp. | | Descriptor: | GLYCEROL, Nucleoside diphosphate kinase B, [[(2R,3R,4R,5R)-5-(2-azanyl-6-oxidanylidene-1H-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methoxy-sulfanyl-phosphoryl] dihydrogen phosphate | | Authors: | Feracci, M, Chazot, A. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | An exonuclease-resistant chain-terminating nucleotide analogue targeting the SARS-CoV-2 replicase complex.
Nucleic Acids Res., 52, 2024
|
|
8PYQ
 
 | | 10 micrometer HEWL crystals solved at room-temperature using fixed-target serial crystallography. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Mason, T.J, Carrillo, M, Beale, J.H, Padeste, C. | | Deposit date: | 2023-07-25 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Micro-structured polymer fixed targets for serial crystallography at synchrotrons and XFELs.
Iucrj, 10, 2023
|
|
8PYO
 
 | | 5 micrometer HEWL crystals solved at room-temperature using fixed-target serial crystallography. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Mason, T.J, Carrillo, M, Beale, J.H, Padeste, C. | | Deposit date: | 2023-07-25 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Micro-structured polymer fixed targets for serial crystallography at synchrotrons and XFELs.
Iucrj, 10, 2023
|
|
8PYP
 
 | | 25 micrometer HEWL crystals solved at room-temperature using fixed-target serial crystallography. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Mason, T.J, Carrillo, M, Beale, J.H, Padeste, C. | | Deposit date: | 2023-07-25 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Micro-structured polymer fixed targets for serial crystallography at synchrotrons and XFELs.
Iucrj, 10, 2023
|
|
8TKM
 
 | | Murine NF-kappaB p50 Rel Homology Region homodimer in complex with 17-mer kappaB DNA from human interleukin-6 (IL-6) promoter | | Descriptor: | 17-mer kappaB DNA, Nuclear factor NF-kappa-B p50 subunit | | Authors: | Zhu, N, Mealka, M, Mitchel, S, Rogers, W.E, Huxford, T. | | Deposit date: | 2023-07-25 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallographic Study of Preferred Spacing by the NF-kappa B p50 Homodimer on kappa B DNA.
Biomolecules, 13, 2023
|
|
8TK8
 
 | | Human Type 3 IP3 Receptor - Resting State (+IP3/ATP) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
8TKG
 
 | | Human Type 3 IP3 Receptor - Resting State (+IP3/ATP) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
8TKF
 
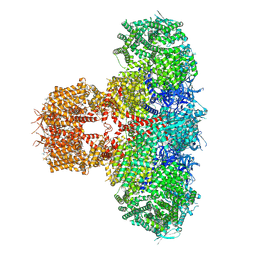 | | Human Type 3 IP3 Receptor - Activated State (+IP3/ATP/JD Ca2+) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
