6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
6D5Y
 
 | | Crystal structure of ERK2 G169D mutant | | Descriptor: | Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Jaiswal, B.S, Wang, W. | | Deposit date: | 2018-04-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | ERK Mutations and Amplification Confer Resistance to ERK-Inhibitor Therapy.
Clin. Cancer Res., 24, 2018
|
|
6CPW
 
 | | Discovery of 3(S)-thiomethyl pyrrolidine ERK inhibitors for oncology | | Descriptor: | (3S)-N-[3-(4-fluorophenyl)-1H-indazol-5-yl]-3-(methylsulfanyl)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Hruza, A, Hruza, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of 3(S)-thiomethyl pyrrolidine ERK inhibitors for oncology.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6BHT
 
 | | HIV-1 CA hexamer in complex with IP6, orthorhombic crystal form | | Descriptor: | Capsid protein p24, INOSITOL HEXAKISPHOSPHATE | | Authors: | Zadrozny, K, Wagner, J.M, Ganser-Pornillos, B.K, Pornillos, O. | | Deposit date: | 2017-10-31 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Inositol phosphates are assembly co-factors for HIV-1.
Nature, 560, 2018
|
|
6BHS
 
 | | HIV-1 CA hexamer in complex with IP6, hexagonal crystal form | | Descriptor: | Capsid protein p24, INOSITOL HEXAKISPHOSPHATE | | Authors: | Zadrozny, K, Wagner, J.M, Ganser-Pornillos, B.K, Pornillos, O. | | Deposit date: | 2017-10-31 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Inositol phosphates are assembly co-factors for HIV-1.
Nature, 560, 2018
|
|
6BCX
 
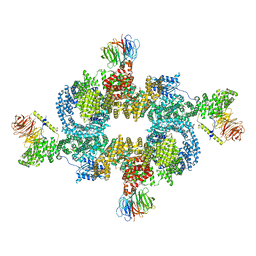 | | mTORC1 structure refined to 3.0 angstroms | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E-binding protein 1, MAGNESIUM ION, ... | | Authors: | Pavletich, N.P, Yang, H. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40.
Nature, 552, 2017
|
|
6BCU
 
 | |
6B2K
 
 | | E45A/R132T mutant of HIV-1 capsid protein | | Descriptor: | CHLORIDE ION, HIV-1 capsid protein, IODIDE ION | | Authors: | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-09-20 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization.
Nat Commun, 14, 2023
|
|
6B2J
 
 | |
6B2I
 
 | | E45A mutant of the HIV-1 capsid protein | | Descriptor: | CHLORIDE ION, HIV-1 capsid protein, IODIDE ION | | Authors: | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-09-20 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization.
Nat Commun, 14, 2023
|
|
6B2H
 
 | |
6B2G
 
 | | P38A mutant of HIV-1 capsid protein | | Descriptor: | CHLORIDE ION, HIV-1 capsid protein, IODIDE ION | | Authors: | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-09-20 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization.
Nat Commun, 14, 2023
|
|
6AYA
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Nup153 peptide | | Descriptor: | CHLORIDE ION, HIV-1 capsid protein, IODIDE ION, ... | | Authors: | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization.
Nat Commun, 14, 2023
|
|
6AY9
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with CPSF6 peptide | | Descriptor: | CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 6, HIV-1 capsid protein, ... | | Authors: | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization.
Nat Commun, 14, 2023
|
|
6AXY
 
 | |
6AXX
 
 | |
6AXW
 
 | |
6AXV
 
 | |
6AXT
 
 | |
6AXS
 
 | |
6AXR
 
 | |
6ATE
 
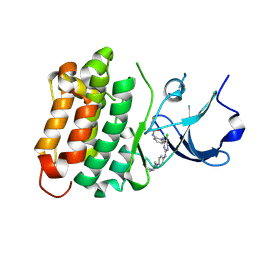 | | SRC kinase bound to covalent inhibitor | | Descriptor: | N-{2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]phenyl}propanamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2017-08-28 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.
Cell Chem Biol, 26, 2019
|
|
6ANL
 
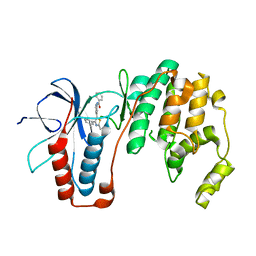 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[1,2-b]pyridazine-based p38 MAP Kinase Inhibitors | | Descriptor: | Mitogen-activated protein kinase 14, TAK-715 | | Authors: | Snell, G.P, Okada, K, Bragstad, K, Sang, B.-C. | | Deposit date: | 2017-08-14 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of imidazo[1,2-b]pyridazine-based p38 MAP kinase inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
5YWC
 
 | |
5YWA
 
 | |
