3WJG
 
 | | Crystal structure of mutant nitrobindin M75L/H76L/Q96C/M148W/H158L (NB10) from Arabidopsis thaliana | | Descriptor: | GLYCEROL, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Mizohata, E, Fukumoto, K, Onoda, A, Bocola, M, Arlt, M, Inoue, T, Schwaneberg, U, Hayashi, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A Rhodium Complex-linked Hybrid Biocatalyst: Stereo-controlled Phenylacetylene Polymerization within an Engineered Protein Cavity
CHEMCATCHEM, 2014
|
|
3WD0
 
 | | Serratia marcescens Chitinase B, tetragonal form | | Descriptor: | Chitinase B, DITHIANE DIOL, GLYCEROL, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4L01
 
 | |
4L00
 
 | |
4NV5
 
 | | C50A mutant of Synechococcus VKOR, C2 crystal form (dehydrated) | | Descriptor: | UBIQUINONE-10, VKORC1/thioredoxin domain protein | | Authors: | Liu, S, Cheng, W, Fowle Grider, R, Shen, G, Li, W. | | Deposit date: | 2013-12-04 | | Release date: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structures of an intramembrane vitamin K epoxide reductase homolog reveal control mechanisms for electron transfer.
Nat Commun, 5, 2014
|
|
4NV2
 
 | | C50A mutant of Synechococcus VKOR, C2221 crystal form | | Descriptor: | UBIQUINONE-10, VKORC1/thioredoxin domain protein | | Authors: | Liu, S, Cheng, W, Fowle Grider, R, Shen, G, Li, W. | | Deposit date: | 2013-12-04 | | Release date: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Structures of an intramembrane vitamin K epoxide reductase homolog reveal control mechanisms for electron transfer.
Nat Commun, 5, 2014
|
|
4G2K
 
 | | Crystal structure of the Marburg Virus GP2 ectodomain in its post-fusion conformation | | Descriptor: | CHLORIDE ION, GLYCEROL, General control protein GCN4, ... | | Authors: | Malashkevich, V.N, Koellhoffer, J.F, Harrison, J.S, Toro, R, Bhosle, R.C, Chandran, K, Lai, J.R, Almo, S.C. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Marburg Virus GP2 Core Domain in Its Postfusion Conformation.
Biochemistry, 51, 2012
|
|
4HI3
 
 | |
4HU5
 
 | |
4HSX
 
 | | Structure of the L100F mutant of dehaloperoxidase-hemoglobin A from Amphitrite ornata with 4-bromophenol | | Descriptor: | 4-BROMOPHENOL, Dehaloperoxidase A, GLYCEROL, ... | | Authors: | Thompson, M.K, Plummer, A, Franzen, S. | | Deposit date: | 2012-10-31 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Role of polarity of the distal pocket in the control of inhibitor binding in dehaloperoxidase-hemoglobin.
Biochemistry, 52, 2013
|
|
4HSW
 
 | | Structure of the L100F mutant of dehaloperoxidase-hemoglobin A from Amphitrite ornata | | Descriptor: | Dehaloperoxidase A, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Thompson, M.K, Plummer, A, Franzen, S. | | Deposit date: | 2012-10-31 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Role of polarity of the distal pocket in the control of inhibitor binding in dehaloperoxidase-hemoglobin.
Biochemistry, 52, 2013
|
|
4HTM
 
 | | Mechanism of CREB Recognition and Coactivation by the CREB Regulated Transcriptional Coactivator CRTC2 | | Descriptor: | CREB-regulated transcription coactivator 2, ZINC ION | | Authors: | Luo, Q, Viste, K, Urday-Zaa, J.C, Kumar, G.S, Tsai, W.-W, Talai, A, Mayo, K, Montminy, M, Radhakrishnan, I. | | Deposit date: | 2012-11-01 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein kinase N1, a cell inhibitor of Akt kinase, has a central role in quality control of germinal center formation
Proc.Natl.Acad.Sci.USA, 2012
|
|
7QHS
 
 | | S. cerevisiae CMGE nucleating origin DNA melting | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, ... | | Authors: | Lewis, J.S, Sousa, J.S, Costa, A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of replication origin melting nucleated by CMG helicase assembly.
Nature, 606, 2022
|
|
5OY5
 
 | |
5VYG
 
 | | Crystal structure of hFA9 EGF repeat with O-glucose trisaccharide | | Descriptor: | CALCIUM ION, Coagulation factor IX, alpha-D-xylopyranose-(1-3)-alpha-D-xylopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Yu, H.J, Li, H.L. | | Deposit date: | 2017-05-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | O-Glycosylation modulates the stability of epidermal growth factor-like repeats and thereby regulates Notch trafficking
J. Biol. Chem., 292, 2017
|
|
6CK3
 
 | | Co-crytsal Structure of MNK2 in Complex With an Inhibitor | | Descriptor: | (3R)-3-methyl-5-[(pyrimidin-4-yl)amino]-2,3-dihydro-1H-isoindol-1-one, CHLORIDE ION, MAP kinase-interacting serine/threonine-protein kinase 2, ... | | Authors: | Han, Q. | | Deposit date: | 2018-02-27 | | Release date: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6CJW
 
 | | Crystal Structure of Mnk2-D228G in Complex With Inhibitor | | Descriptor: | 3-(pyridin-4-yl)imidazo[1,2-b]pyridazine, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-27 | | Release date: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6CJ5
 
 | | Crystal Structure of Mnk2-D228G in Complex With Inhibitor | | Descriptor: | 3-(pyridin-3-yl)imidazo[1,2-a]pyridine-8-carboxamide, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6CKI
 
 | | Co-crystal structure of MNK2 in Complex With Inhibitor | | Descriptor: | 3,3-dimethyl-6-[(pyrimidin-4-yl)amino]-2,3-dihydroimidazo[1,5-a]pyridine-1,5-dione, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6CJH
 
 | | Co-crystal structure of MNK2 in complex with an inhibitor | | Descriptor: | 3-phenyl-5-(pyridin-4-yl)-1H-indazole, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6CJE
 
 | | Crystal Structure of Mnk2-D228G in complex with Inhibitor | | Descriptor: | 4-[(9H-purin-6-yl)amino]benzamide, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
3UM7
 
 | |
4YWK
 
 | |
4YWL
 
 | |
5W43
 
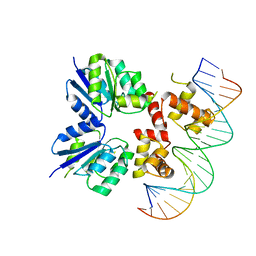 | | Structure of the two-component response regulator RcsB-DNA complex | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*CP*TP*AP*AP*GP*AP*TP*TP*TP*TP*TP*CP*CP*TP*AP*AP*AP*TP*C)-3'), Transcriptional regulatory protein RcsB | | Authors: | Filippova, E.V, Warwzak, Z, Pshenychnyi, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-06-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural Basis for DNA Recognition by the Two-Component Response Regulator RcsB.
MBio, 9, 2018
|
|
