5DB5
 
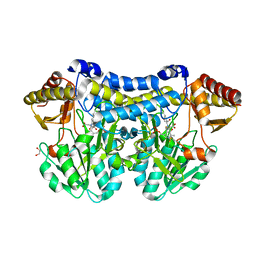 | | Crystal structure of PLP-bound E. coli SufS (cysteine persulfide intermediate) in space group P21 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, CYSTEINE, ... | | Authors: | Arbing, M.A, Shin, A, Koo, C.W, Medrano-Soto, A, Eisenberg, D. | | Deposit date: | 2015-08-20 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of PLP-bound E. coli SufS (cysteine persulfide intermediate) in space group P21
To Be Published
|
|
5OGZ
 
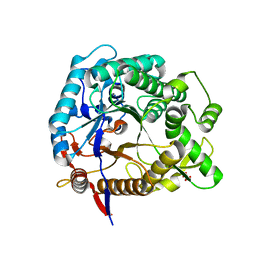 | |
9QLE
 
 | | Human myoferlin (1-1997) in complex with an MSP2N2 lipid nanodisc (15 mol% DOPS, 2 mol% PI(4,5)P2) | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, CALCIUM ION, Myoferlin | | Authors: | Cretu, C, Moser, T. | | Deposit date: | 2025-03-20 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into lipid membrane binding by human ferlins.
Embo J., 44, 2025
|
|
6TN9
 
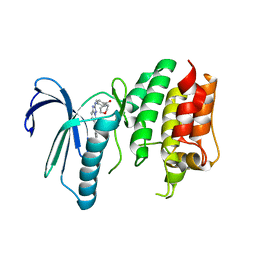 | | X-RAY STRUCTURE OF MPS1 IN COMPLEX WITH COMPOUND 16 | | Descriptor: | Dual specificity protein kinase TTK, [4-[[6-(3,5-dimethyl-4-oxidanyl-phenyl)-[1,2,4]triazolo[1,5-a]pyridin-2-yl]amino]phenyl]-morpholin-4-yl-methanone | | Authors: | Marquardt, T, Holton, S.J, Schulze, V.K, Klar, U, Kosemund, D, Siemeister, G, Bader, B, Prechtl, S, Briem, H, Schirok, H, Bohlmann, R, Nguyen, D, Fernandez-Montalvan, A, Boemer, U, Eberspaecher, U, Brands, M, Nussbaum, F, Koppitz, M. | | Deposit date: | 2019-12-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Treating Cancer by Spindle Assembly Checkpoint Abrogation: Discovery of Two Clinical Candidates, BAY 1161909 and BAY 1217389, Targeting MPS1 Kinase.
J.Med.Chem., 63, 2020
|
|
5M1E
 
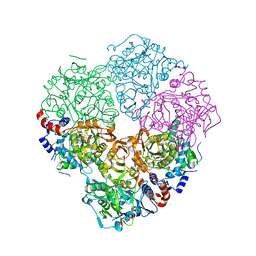 | | Crystal structure of N-terminally tagged UbiD from E. coli reconstituted with prFMN cofactor | | Descriptor: | (16~{R})-11,12,14,14-tetramethyl-3,5-bis(oxidanylidene)-8-[(2~{S},3~{S},4~{R})-2,3,4-tris(oxidanyl)-5-phosphonooxy-pentyl]-1,4,6,8-tetrazatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-2(7),9(17),10,12-tetraene-16-sulfonic acid, 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, MANGANESE (II) ION, ... | | Authors: | Marshall, S.A, Leys, D. | | Deposit date: | 2016-10-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Oxidative Maturation and Structural Characterization of Prenylated FMN Binding by UbiD, a Decarboxylase Involved in Bacterial Ubiquinone Biosynthesis.
J. Biol. Chem., 292, 2017
|
|
6S8V
 
 | | Structure of the high affinity Anticalin P3D11 in complex with the human CD98 heavy chain ectodomain | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, Neutrophil gelatinase-associated lipocalin | | Authors: | Schiefner, A, Deuschle, F.-C, Skerra, A. | | Deposit date: | 2019-07-10 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of a high affinity Anticalin®directed against human CD98hc for theranostic applications.
Theranostics, 10, 2020
|
|
4LTA
 
 | |
6IBX
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 5 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-[(3~{S})-1-methylpiperidin-3-yl]pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Pawlik, H, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Synthesis of amide and sulfonamide substituted N-aryl 6-aminoquinoxalines as PFKFB3 inhibitors with improved physicochemical properties.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
7CJT
 
 | | Crystal Structure of SETDB1 Tudor domain in complexed with (R,R)-59 | | Descriptor: | 2-[[(3~{R},5~{R})-1-methyl-5-(4-phenylmethoxyphenyl)piperidin-3-yl]amino]-3-prop-2-enyl-5~{H}-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Guo, Y.P, Liang, X, Mao, X, Wu, C, Luyi, H, Yang, S. | | Deposit date: | 2020-07-13 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.474 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5O4V
 
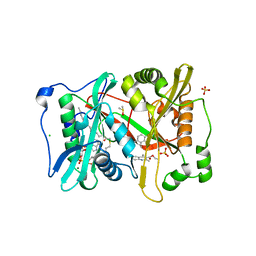 | | P.vivax NMT with aminomethylindazole and quinoline inhibitors bound | | Descriptor: | 1-[5-(4-fluoranyl-2-methyl-phenyl)-1~{H}-indazol-3-yl]-~{N},~{N}-dimethyl-methanamine, 2-oxopentadecyl-CoA, CHLORIDE ION, ... | | Authors: | Brannigan, J.A, Wilkinson, A.J. | | Deposit date: | 2017-05-31 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-derived inhibitors of human N-myristoyltransferase block capsid assembly and replication of the common cold virus.
Nat Chem, 10, 2018
|
|
6FVF
 
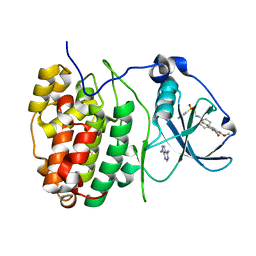 | | The Structure of CK2alpha with CCh503 bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, [1-[2-(phenylsulfonylamino)ethyl]piperidin-4-yl]methyl 5-fluoranyl-2-methoxy-1~{H}-indole-3-carboxylate | | Authors: | Brear, P, Prudent, R, Laudet, B, Filhol, O, Cochet, C, Sautel, C, Moucadel, V, Bestgen, B, Engel, M, Ettaoussi, M, Lomberget, T, Le Borgne, M, Kufareva, I, Abagyan, R, Hyvonen, M. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of holoenzyme-disrupting chemicals as substrate-selective CK2 inhibitors.
Sci Rep, 9, 2019
|
|
7KQ6
 
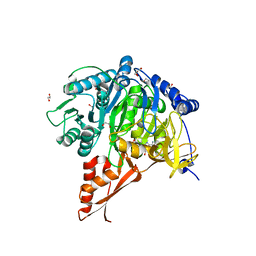 | |
5IHA
 
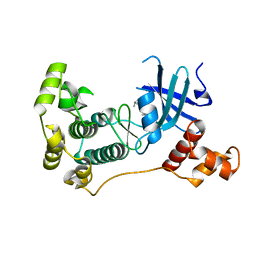 | | MELK in complex with NVS-MELK8F | | Descriptor: | 1-methyl-4-(4-{4-[3-(2-methylpropoxy)pyridin-4-yl]-1H-pyrazol-1-yl}phenyl)piperazine, Maternal embryonic leucine zipper kinase | | Authors: | Sprague, E.R, Brazell, T. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Toward the Validation of Maternal Embryonic Leucine Zipper Kinase: Discovery, Optimization of Highly Potent and Selective Inhibitors, and Preliminary Biology Insight.
J.Med.Chem., 59, 2016
|
|
4EMV
 
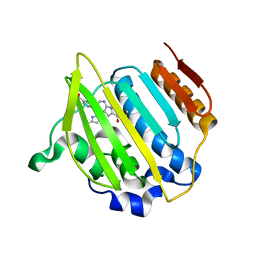 | | Crystal structure of a topoisomerase ATP inhibitor | | Descriptor: | 5-{2-(ethylcarbamoyl)-4-[3-(trifluoromethyl)-1H-pyrazol-1-yl]-1H-pyrrolo[2,3-b]pyridin-5-yl}pyridine-3-carboxylic acid, DNA topoisomerase IV, B subunit | | Authors: | Boriack-Sjodin, P.A, Manchester, J, Hull, K. | | Deposit date: | 2012-04-12 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a novel azaindole class of antibacterial agents targeting the ATPase domains of DNA gyrase and Topoisomerase IV.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5EST
 
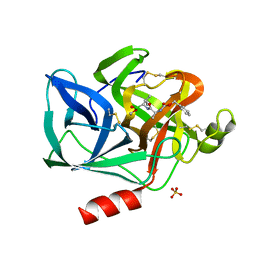 | | Crystallographic analysis of the inhibition of porcine pancreatic elastase by a peptidyl boronic acid: structure of a reaction intermediate | | Descriptor: | CALCIUM ION, ELASTASE, N~2~-[(benzyloxy)carbonyl]-N-[(1R,2S)-1-(dihydroxyboranyl)-2-methylbutyl]-L-alaninamide, ... | | Authors: | Takahashi, L.H, Radhakrishnan, R, Rosenfieldjunior, R.E, Meyerjunior, E.F. | | Deposit date: | 1989-05-15 | | Release date: | 1992-04-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystallographic analysis of the inhibition of porcine pancreatic elastase by a peptidyl boronic acid: structure of a reaction intermediate.
Biochemistry, 28, 1989
|
|
7NIV
 
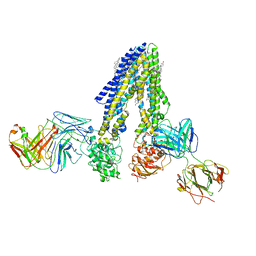 | | Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab and QA2-Fab (phosphatidylcholine-bound, occluded conformation) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4B1 Fab-fragment light chain, CHOLESTEROL, ... | | Authors: | Nosol, K, Locher, K.P. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of ABCB4 provide insight into phosphatidylcholine translocation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NIW
 
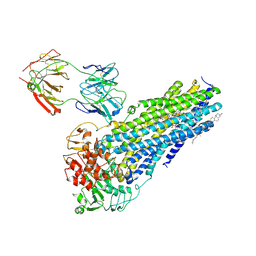 | | Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab (posaconazole-bound, inward-open conformation) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4B1 Fab-fragment heavy chain, 4B1 Fab-fragment light chain, ... | | Authors: | Nosol, K, Locher, K.P. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of ABCB4 provide insight into phosphatidylcholine translocation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5F13
 
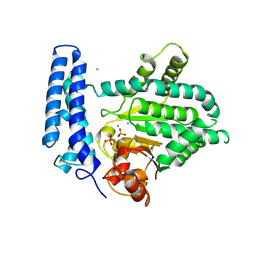 | | Structure of Mn bound DUF89 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Nocek, B, Skarina, T, Joachimiak, A, Savchenko, A, Yakunin, A. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | A family of metal-dependent phosphatases implicated in metabolite damage-control.
Nat.Chem.Biol., 12, 2016
|
|
5AXV
 
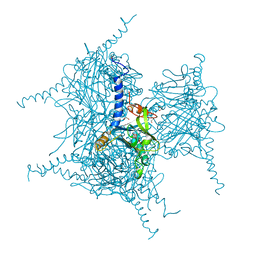 | | Crystal Structure of Cypovirus Polyhedra R13K Mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Abe, S, Ijiri, H, Negishi, H, Yamanaka, H, Sasaki, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2015-08-01 | | Release date: | 2015-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Design of Enzyme-Encapsulated Protein Containers by In Vivo Crystal Engineering
Adv. Mater. Weinheim, 27, 2015
|
|
3IDG
 
 | |
3GNM
 
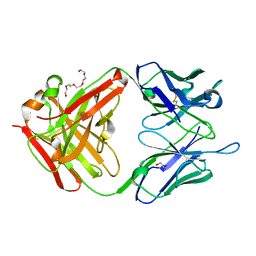 | | The crystal structure of the JAA-F11 monoclonal antibody Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, JAA-F11 Fab Antibody Fragment, Heavy Chain, ... | | Authors: | Gulick, A.M, Rittenhouse-Olson, K, Jadey, S. | | Deposit date: | 2009-03-17 | | Release date: | 2010-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computational Screening of the Human TF-Glycome Provides a Structural Definition for the Specificity of Anti-Tumor Antibody JAA-F11.
Plos One, 8, 2013
|
|
5A61
 
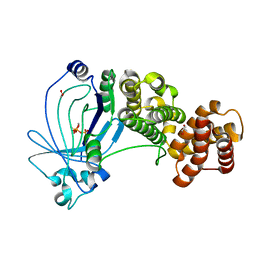 | | Crystal structure of full-length E. coli ygiF in complex with tripolyphosphate and two manganese ions. | | Descriptor: | 1,2-ETHANEDIOL, INORGANIC TRIPHOSPHATASE, MANGANESE (II) ION, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5A5Y
 
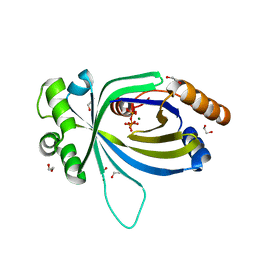 | | Crystal structure of AtTTM3 in complex with tripolyphosphate and magnesium ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, TRIPHOSPHATE, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5IS7
 
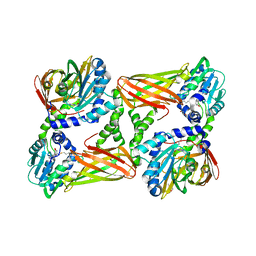 | | Crystal structure of mouse CARM1 in complex with decarboxylated SAH | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-(3-aminopropyl)-5'-thioadenosine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with decarboxylated SAH
To Be Published
|
|
3MNQ
 
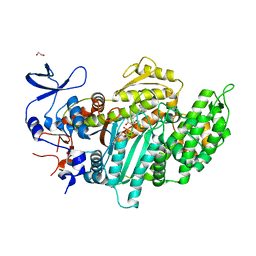 | | Crystal structure of myosin-2 motor domain in complex with ADP-metavanadate and resveratrol | | Descriptor: | 1,2-ETHANEDIOL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Schneider, J, Taft, M, Backhaus, A, Baruch, P, Fedorov, R, Manstein, D.J. | | Deposit date: | 2010-04-22 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of resveratrol regulation of myosin activity.
To be Published
|
|
