6QTA
 
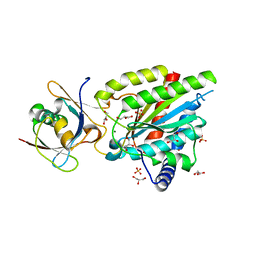 | | Crystal structure of Rea1-MIDAS/Rsa4-UBL complex from Chaetomium thermophilum | | Descriptor: | GLYCEROL, MAGNESIUM ION, Midasin,Midasin, ... | | Authors: | Ahmed, Y.L, Thoms, M, Hurt, E, Sinning, I. | | Deposit date: | 2019-02-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of Rea1-MIDAS bound to its ribosome assembly factor ligands resembling integrin-ligand-type complexes.
Nat Commun, 10, 2019
|
|
7S7L
 
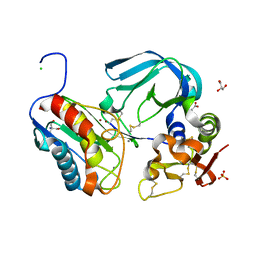 | | Complex of tissue inhibitor of metalloproteinases-1 (TIMP-1) mutant (L34G/M66S/E67Y/L133N/S155L) with matrix metalloproteinase-3 catalytic domain (MMP-3cd) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Coban, M, Raeeszadeh-Sarmazdeh, M, Hockla, A, Sankaran, B, Radisky, E.S. | | Deposit date: | 2021-09-16 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Engineering of tissue inhibitor of metalloproteinases TIMP-1 for fine discrimination between closely related stromelysins MMP-3 and MMP-10.
J.Biol.Chem., 298, 2022
|
|
6XT3
 
 | |
6QRF
 
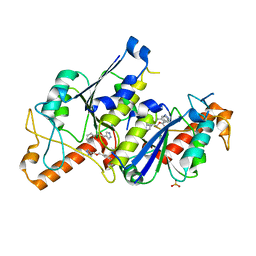 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | (phenylmethyl) 1~{H}-pyrazole-4-carboxylate, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
6DJW
 
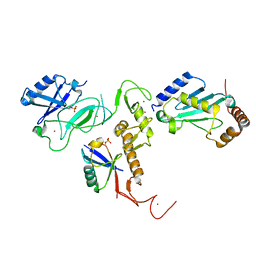 | | Crystal Structure of pParkin (REP and RING2 deleted)-pUb-UbcH7 complex | | Descriptor: | RBR-type E3 ubiquitin transferase,RBR-type E3 ubiquitin transferase, Ubiquitin, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Sauve, V, Sung, G, Trempe, J.F, Gehring, K. | | Deposit date: | 2018-05-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Mechanism of parkin activation by phosphorylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8U57
 
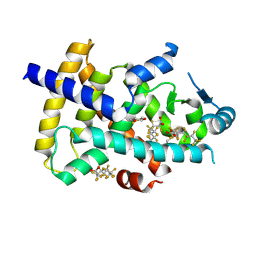 | | PPARg LBD in complex with perfluorooctanoic acid (PFOA) | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Peroxisome proliferator-activated receptor gamma, pentadecafluorooctanoic acid | | Authors: | Pederick, J.L, Frkic, R.L, McDougal, D.P, Bruning, J.B. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural basis for the activation of peroxisome proliferator-activated receptor gamma (PPAR gamma ) by perfluorooctanoic acid (PFOA).
Chemosphere, 354, 2024
|
|
7SBH
 
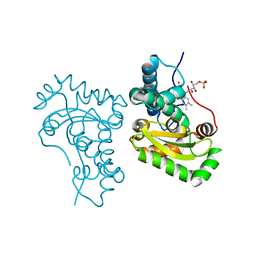 | | Crystal structure of the iron superoxide dismutase from Acinetobacter sp. Ver3 | | Descriptor: | FE (III) ION, FLAVIN MONONUCLEOTIDE, Superoxide dismutase | | Authors: | Steimbruch, B.A, Albanesi, D, Repizo, G.D, Lisa, M.N. | | Deposit date: | 2021-09-24 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The distinctive roles played by the superoxide dismutases of the extremophile Acinetobacter sp. Ver3.
Sci Rep, 12, 2022
|
|
6QTS
 
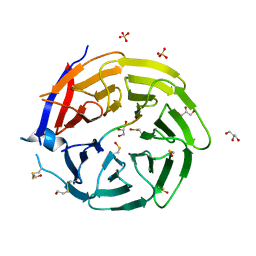 | |
7S58
 
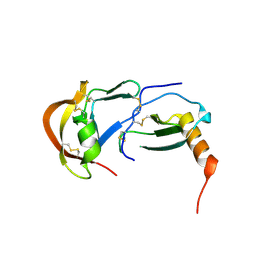 | | Crystal Structure of the tick evasin EVA-P974 complexed to human chemokine CCL7 | | Descriptor: | C-C motif chemokine 7, Evasin P974 | | Authors: | Bhusal, R.P, Devkota, S.R, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-guided engineering of tick evasins for targeting chemokines in inflammatory diseases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6QRU
 
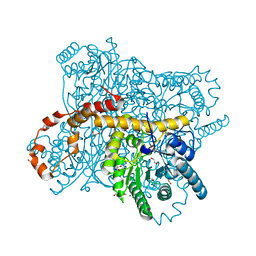 | | X-ray radiation dose series on xylose isomerase - 2.01 MGy | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Taberman, H, Bury, C.S, van der Woerd, M.J, Snell, E.H, Garman, E.F. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural knowledge or X-ray damage? A case study on xylose isomerase illustrating both.
J.Synchrotron Radiat., 26, 2019
|
|
6JTO
 
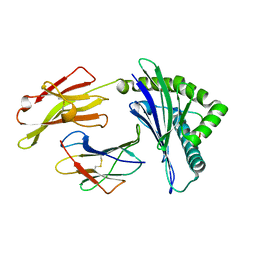 | | Crystal structure of HLA-C05 in complex with a tumor mut10m peptide | | Descriptor: | 10-mer peptide, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Bai, P, Zhou, Q, Wei, P, Lei, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-04-15 | | Last modified: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational discovery of a cancer neoepitope harboring the KRAS G12D driver mutation.
Sci China Life Sci, 2021
|
|
6GAR
 
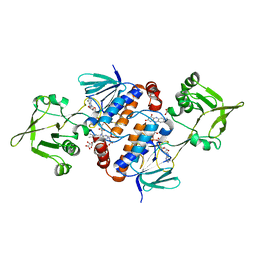 | | Crystal structure of oxidised ferredoxin/flavodoxin NADP+ oxidoreductase 1 (FNR1) from Bacillus cereus | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Skramo, S, Gudim, I, Hersleth, H.-P. | | Deposit date: | 2018-04-12 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Characterization of Different Flavodoxin Reductase-Flavodoxin (FNR-Fld) Interactions Reveals an Efficient FNR-Fld Redox Pair and Identifies a Novel FNR Subclass.
Biochemistry, 57, 2018
|
|
6QU0
 
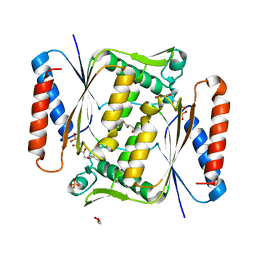 | | Structure of azoreductase from Bacillus sp. A01 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase, GLYCEROL, ... | | Authors: | Savino, S, Fraaije, M.W. | | Deposit date: | 2019-02-26 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic and Crystallographic Studies of Azoreductase AzoA fromBacillus wakoensisA01.
Acs Chem.Biol., 15, 2020
|
|
6DKK
 
 | | Structure of BoNT | | Descriptor: | Botulinum neurotoxin type A, PHOSPHATE ION | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2018-05-29 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A viral-fusion-peptide-like molecular switch drives membrane insertion of botulinum neurotoxin A1.
Nat Commun, 9, 2018
|
|
6GG4
 
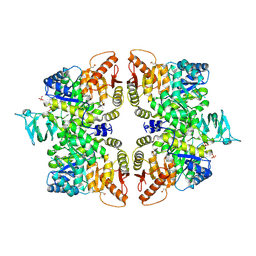 | | Crystal structure of M2 PYK in complex with Phenyalanine. | | Descriptor: | PHENYLALANINE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | McNae, I.W, Yuan, M, Walkinshaw, M.D. | | Deposit date: | 2018-05-02 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | An allostatic mechanism for M2 pyruvate kinase as an amino-acid sensor.
Biochem. J., 475, 2018
|
|
6DKV
 
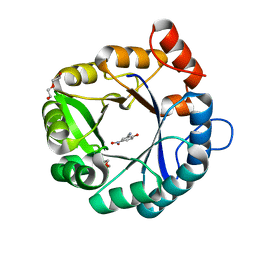 | |
6GH2
 
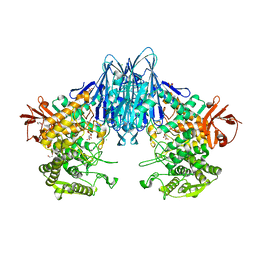 | | Paenibacillus sp. YM1 laminaribiose phosphorylase with alpha-glc-1-phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Kuhaudomlarp, S, Walpole, S, Stevenson, C.E.M, Nepogodiev, S.A, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2018-05-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unravelling the Specificity of Laminaribiose Phosphorylase from Paenibacillus sp. YM-1 towards Donor Substrates Glucose/Mannose 1-Phosphate by Using X-ray Crystallography and Saturation Transfer Difference NMR Spectroscopy.
Chembiochem, 20, 2019
|
|
6QT1
 
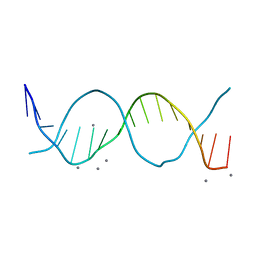 | | Radiation damage study on a 16mer DNA segment, structure at 0.48 MGy dose | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*GP*AP*AP*AP*TP*TP*TP*CP*CP*AP*GP*C)-3') | | Authors: | Bugris, V, Harmat, V, Ferenc, G, Brockhauser, S, Carmichael, I, Garman, E.F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Radiation-damage investigation of a DNA 16-mer.
J.Synchrotron Radiat., 26, 2019
|
|
7S7M
 
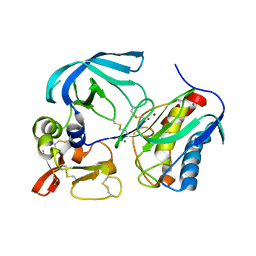 | | Complex of tissue inhibitor of metalloproteinases-1 (TIMP-1) mutant (L34G/M66D/T98G/P131S/Q153N) with matrix metalloproteinase-3 catalytic domain (MMP-3cd) | | Descriptor: | CALCIUM ION, Metalloproteinase inhibitor 1, Stromelysin-1, ... | | Authors: | Coban, M, Raeeszadeh-Sarmazdeh, M, Sankaran, B, Hockla, A, Radisky, E.S. | | Deposit date: | 2021-09-16 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineering of tissue inhibitor of metalloproteinases TIMP-1 for fine discrimination between closely related stromelysins MMP-3 and MMP-10.
J.Biol.Chem., 298, 2022
|
|
6GU2
 
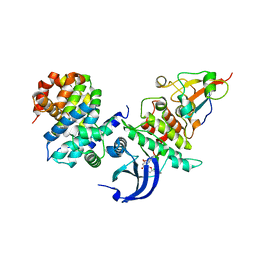 | | CDK1/CyclinB/Cks2 in complex with Flavopiridol | | Descriptor: | 2-(2-chlorophenyl)-8-[(3~{R},4~{R})-1-methyl-3-oxidanyl-piperidin-4-yl]-5,7-bis(oxidanyl)chromen-4-one, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GHW
 
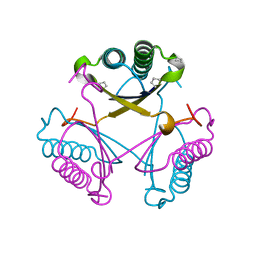 | | Substituting the prolines of 4-oxalocrotonate tautomerase with non-canonical analogue (2S)-3,4-dehydroproline | | Descriptor: | 2-hydroxymuconate tautomerase, CALCIUM ION | | Authors: | Pavkov-Keller, T, Lukesch, M.S, Wiltschi, B, Gruber, K. | | Deposit date: | 2018-05-09 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substituting the catalytic proline of 4-oxalocrotonate tautomerase with non-canonical analogues reveals a finely tuned catalytic system.
Sci Rep, 9, 2019
|
|
8U7J
 
 | | Crystal Structure of Staphylococcus aureus PLP synthase complex | | Descriptor: | GLUTAMINE, PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PdxS, ... | | Authors: | Barra, A.L.C, Brognaro, H, Betzel, C, Nascimento, A.S. | | Deposit date: | 2023-09-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure and dynamics of the staphylococcal pyridoxal 5-phosphate synthase complex reveal transient interactions at the enzyme interface.
J.Biol.Chem., 300, 2024
|
|
6DL0
 
 | | Crystal structure of pohlianin C, an orbitide from Jatropha pohliana | | Descriptor: | pohlianin C | | Authors: | Wang, C.K, King, G.J, Ramalho, S.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-11-07 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Synthesis, Racemic X-ray Crystallographic, and Permeability Studies of Bioactive Orbitides from Jatropha Species.
J. Nat. Prod., 81, 2018
|
|
6X6R
 
 | | Crystal structure of C.difficile ribosyltransferase CDTa in complex with pCl-phenylthioDADMeImmA | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-chlorophenyl)sulfanyl]methyl}pyrrolidin-3-ol, ADP-ribosyltransferase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Pozharski, E. | | Deposit date: | 2020-05-29 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of C.difficile ribosyltransferase CDTa in complex with pCl-phenylthioDADMeImmA
To Be Published
|
|
6QT4
 
 | | Radiation damage study on a 16mer DNA segment, structure at 17.7 MGy dose | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*GP*AP*AP*AP*TP*TP*TP*CP*CP*AP*GP*C)-3') | | Authors: | Bugris, V, Harmat, V, Ferenc, G, Brockhauser, S, Carmichael, I, Garman, E.F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Radiation-damage investigation of a DNA 16-mer.
J.Synchrotron Radiat., 26, 2019
|
|
