1R9F
 
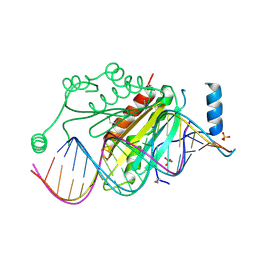 | | Crystal structure of p19 complexed with 19-bp small interfering RNA | | Descriptor: | 5'-R(*CP*GP*UP*AP*CP*GP*CP*GP*GP*AP*AP*UP*AP*CP*UP*UP*CP*GP*AP*UP*U)-3', 5'-R(*UP*CP*GP*AP*AP*GP*UP*AP*UP*UP*CP*CP*GP*CP*GP*UP*AP*CP*GP*UP*U)-3', Core protein P19, ... | | Authors: | Ye, K, Malinina, L, Patel, D.J. | | Deposit date: | 2003-10-28 | | Release date: | 2004-01-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Recognition of small interfering RNA by a viral suppressor of RNA
Nature, 426, 2003
|
|
1RNR
 
 | |
1RBC
 
 | |
1RVJ
 
 | | PHOTOSYNTHETIC REACTION CENTER DOUBLE MUTANT FROM RHODOBACTER SPHAEROIDES WITH ASP L213 REPLACED WITH ASN AND ARG H177 REPLACED WITH HIS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Xu, Q, Axelrod, H.L, Abresch, E.C, Paddock, M.L, Okamura, M.Y, Feher, G. | | Deposit date: | 2003-12-14 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | X-Ray Structure Determination of Three Mutants of the Bacterial Photosynthetic Reaction
Centers from Rb. sphaeroides; Altered Proton Transfer Pathways.
Structure, 12, 2004
|
|
1RWE
 
 | | Enhancing the activity of insulin at receptor edge: crystal structure and photo-cross-linking of A8 analogues | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z, Xu, B, Chu, Y.C, Li, B, Nakagawa, S.H, Qu, Y, Hu, S.Q, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2003-12-16 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enhancing the activity of insulin at the receptor interface: crystal structure and photo-cross-linking of A8 analogues.
Biochemistry, 43, 2004
|
|
1R2F
 
 | |
1R2Y
 
 | | MutM (Fpg) bound to 8-oxoguanine (oxoG) containing DNA | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*TP*CP*CP*AP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | DNA Lesion Recognition by the Bacterial Repair Enzyme MutM.
J.Biol.Chem., 278, 2003
|
|
1R35
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER, TETRAHYDROBIOPTERIN AND 4R-FLUORO-N6-ETHANIMIDOYL-L-LYSINE | | Descriptor: | 4R-FLUORO-N6-ETHANIMIDOYL-L-LYSINE, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Shieh, H.S, Stevens, A.M, Stallings, W.C. | | Deposit date: | 2003-09-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 4-Fluorinated L-lysine analogs as selective i-NOS inhibitors: methodology for introducing fluorine into the lysine side chain.
Org.Biomol.Chem., 1, 2003
|
|
1RGB
 
 | | Phospholipase A2 from Vipera ammodytes meridionalis | | Descriptor: | (9E)-OCTADEC-9-ENAMIDE, Phospholipase A2 | | Authors: | Georgieva, D.N. | | Deposit date: | 2003-11-12 | | Release date: | 2005-01-18 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Asp49 phospholipase A(2)-elaidoylamide complex: a new mode of inhibition.
Biochem.Biophys.Res.Commun., 319, 2004
|
|
1RNV
 
 | | REFINEMENT OF THE CRYSTAL STRUCTURE OF RIBONUCLEASE S. COMPARISON WITH AND BETWEEN THE VARIOUS RIBONUCLEASE A STRUCTURES | | Descriptor: | RIBONUCLEASE S, SULFATE ION | | Authors: | Kim, E.E, Varadarajan, R, Wyckoff, H.W, Richards, F.M. | | Deposit date: | 1992-02-19 | | Release date: | 1994-01-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refinement of the crystal structure of ribonuclease S. Comparison with and between the various ribonuclease A structures.
Biochemistry, 31, 1992
|
|
1RDY
 
 | | T-STATE STRUCTURE OF THE ARG 243 TO ALA MUTANT OF PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Stec, B, Abraham, R, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-05-17 | | Release date: | 1997-01-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the active site mutant (Arg-243-->Ala) in the T and R allosteric states of pig kidney fructose-1,6-bisphosphatase expressed in Escherichia coli.
Protein Sci., 5, 1996
|
|
1REE
 
 | | ENDO-1,4-BETA-XYLANASE II COMPLEX WITH 3,4-EPOXYBUTYL-BETA-D-XYLOSIDE | | Descriptor: | (3S)-3-hydroxybutyl beta-D-xylopyranoside, BENZOIC ACID, ENDO-1,4-BETA-XYLANASE II | | Authors: | Rouvinen, J, Havukainen, R, Torronen, A. | | Deposit date: | 1995-12-21 | | Release date: | 1997-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Covalent binding of three epoxyalkyl xylosides to the active site of endo-1,4-xylanase II from Trichoderma reesei.
Biochemistry, 35, 1996
|
|
1RRA
 
 | | RIBONUCLEASE A FROM RATTUS NORVEGICUS (COMMON RAT) | | Descriptor: | PHOSPHATE ION, PROTEIN (RIBONUCLEASE) | | Authors: | Gupta, V, Muyldermans, S, Wyns, L, Salunke, D. | | Deposit date: | 1998-12-04 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of recombinant rat pancreatic RNase A.
Proteins, 35, 1999
|
|
1RVE
 
 | |
1RN1
 
 | | THREE-DIMENSIONAL STRUCTURE OF GLN 25-RIBONUCLEASE T1 AT 1.84 ANGSTROMS RESOLUTION: STRUCTURAL VARIATIONS AT THE BASE RECOGNITION AND CATALYTIC SITES | | Descriptor: | RIBONUCLEASE T1 ISOZYME, SULFATE ION | | Authors: | Arni, R.K, Pal, G.P, Ravichandran, K.G, Tulinsky, A, Walz Junior, F.G, Metcalf, P. | | Deposit date: | 1991-11-22 | | Release date: | 1994-01-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Three-dimensional structure of Gln25-ribonuclease T1 at 1.84-A resolution: structural variations at the base recognition and catalytic sites.
Biochemistry, 31, 1992
|
|
1R39
 
 | | THE STRUCTURE OF P38ALPHA | | Descriptor: | Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Patel, S.B, Cameron, P.M, Frantz-Wattley, B, O'Neill, E, Becker, J.W, Scapin, G. | | Deposit date: | 2003-10-01 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lattice stabilization and enhanced diffraction in human p38 alpha crystals by protein engineering.
Biochim.Biophys.Acta, 1696, 2004
|
|
1RPO
 
 | |
1R4N
 
 | | APPBP1-UBA3-NEDD8, an E1-ubiquitin-like protein complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ubiquitin-like protein NEDD8, ZINC ION, ... | | Authors: | Walden, H, Podgorski, M.S, Holton, J.M, Schulman, B.A. | | Deposit date: | 2003-10-07 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1.
Mol.Cell, 12, 2003
|
|
1U8H
 
 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ALDKWAS | | Descriptor: | ANTIBODY 2F5 (HEAVY CHAIN), ANTIBODY 2F5 (LIGHT CHAIN), GP41 PEPTIDE | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications
J.Virol., 83, 2009
|
|
1U90
 
 | | Crystal structures of Ral-GppNHp and Ral-GDP reveal two novel binding sites that are also present in Ras and Rap | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Ral-A | | Authors: | Nicely, N.I, Kosak, J, de Serrano, V, Mattos, C. | | Deposit date: | 2004-08-09 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Ral-GppNHp and Ral-GDP Reveal Two Binding Sites that Are Also Present in Ras and Rap
Structure, 12, 2004
|
|
1U04
 
 | |
1U1L
 
 | |
1TRI
 
 | |
1TRR
 
 | |
1UGB
 
 | |
