6RZM
 
 | | Galectin-3C in complex with trifluoroaryltriazole monothiogalactoside derivative:5(Iodine) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-(3-iodanylphenyl)sulfanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Thermodynamic studies of halogen-bond interactions in galectin-3
to be published
|
|
6QN6
 
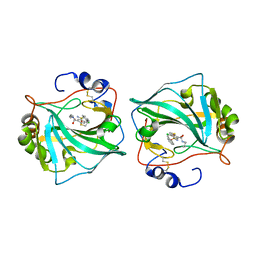 | |
6SYU
 
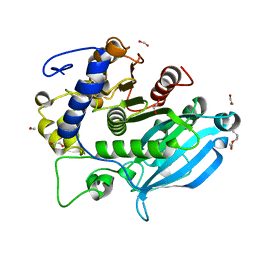 | | The wild type glucuronoyl esterase OtCE15A from Opitutus terrae in complex with xylobiose | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Mazurkewich, S, Navarro Poulsen, J.C, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2019-10-01 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural and biochemical studies of the glucuronoyl esteraseOtCE15A illuminate its interaction with lignocellulosic components.
J.Biol.Chem., 294, 2019
|
|
6RZK
 
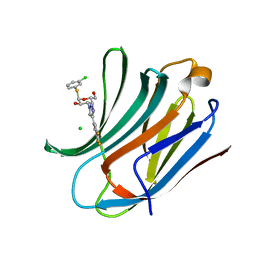 | | Galectin-3C in complex with trifluoroaryltriazole monothiogalactoside derivative:3(Chlorine) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(3-chlorophenyl)sulfanyl-6-(hydroxymethyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.046 Å) | | Cite: | Thermodynamic studies of halogen-bond interactions in galectin-3
to be published
|
|
6RHL
 
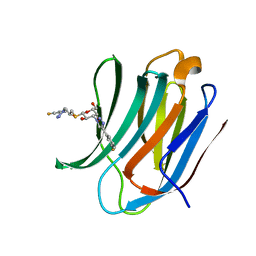 | | Room temperature data of Galectin-3C in complex with a pair of enantiomeric ligands: R enantiomer | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-[(2~{R})-3-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-oxidanyl-propyl]sulfanyl-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-04-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Are crystallographic B-factors suitable for calculating protein conformational entropy?
Phys Chem Chem Phys, 21, 2019
|
|
6T2B
 
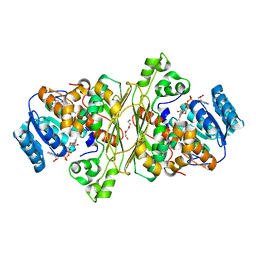 | | Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Glycosyl hydrolase family 109 protein 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Chaberski, E.K, Fredslund, F, Teze, D, Shuoker, B, Kunstmann, S, Karlsson, E.N, Hachem, M.A, Welner, D.H. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Catalytic Acid-Base in GH109 Resides in a Conserved GGHGG Loop and Allows for Comparable alpha-Retaining and beta-Inverting Activity in an N-Acetylgalactosaminidase from Akkermansia muciniphila
Acs Catalysis, 2020
|
|
2V3R
 
 | | Hypocrea jecorina Cel7A in complex with (S)-dihydroxy-phenanthrenolol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(2S)-2-HYDROXY-3-(9-PHENANTHRYLOXY)PROPYL]AMINO}PROPANE-1,3-DIOL, COBALT (II) ION, ... | | Authors: | Fagerstrom, A, Sandgren, M, Berg, U, Stahlberg, J. | | Deposit date: | 2007-06-21 | | Release date: | 2008-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Study of the Chiral Recognition Mechanisms of Cellobiohydrolase Cel7A for Ligands Based on the Beta-Blocker Motif: Crystal Structures, Microcalorimetry and Computational Modelling of Cel7A-Inhibitor Complexes.
To be Published
|
|
2VC4
 
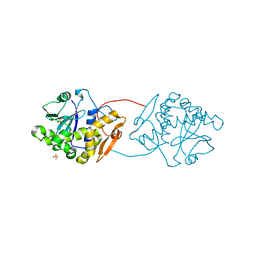 | | Ricin A-Chain (Recombinant) E177D Mutant | | Descriptor: | GLYCEROL, RICIN A CHAIN, SULFATE ION | | Authors: | Marsden, C.J, Fulop, V. | | Deposit date: | 2007-09-18 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The Isolation and Characterisation of Temperature-Dependent Ricin a Chain Molecules in Saccharomyces Cerevisiae
FEBS J., 274, 2007
|
|
2V69
 
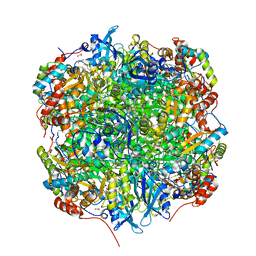 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with a large- subunit mutation D473E | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Satagopan, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2007-07-14 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of Altered Large-Subunit Loop-6-Carboxy-Terminus Interactions that Influence Catalytic Efficiency and Co2-O2 Specificity of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase
Biochemistry, 46, 2007
|
|
2V68
 
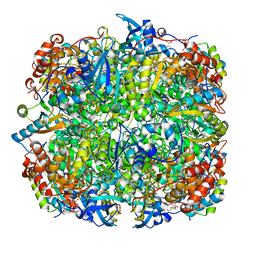 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with large- subunit mutations V331A, T342I | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Satagopan, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2007-07-13 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Altered Large-Subunit Loop-6-Carboxy-Terminus Interactions that Influence Catalytic Efficiency and Co2 O2 Specificity of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase
Biochemistry, 46, 2007
|
|
2VJO
 
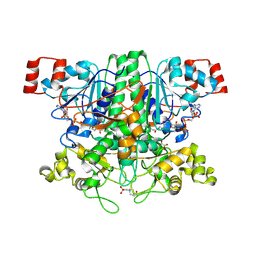 | | Formyl-CoA transferase mutant variant Q17A with aspartyl-CoA thioester intermediates and oxalate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, COENZYME A, ... | | Authors: | Berthold, C.L, Toyota, C.G, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-12-11 | | Release date: | 2007-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reinvestigation of the Catalytic Mechanism of Formyl-Coa Transferase, a Class III Coa-Transferase.
J.Biol.Chem., 283, 2008
|
|
2V3W
 
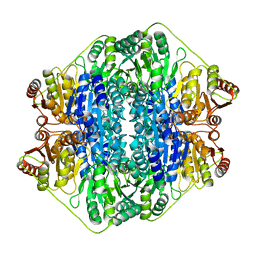 | | Crystal structure of the benzoylformate decarboxylase variant L461A from Pseudomonas putida | | Descriptor: | BENZOYLFORMATE DECARBOXYLASE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Gocke, D, Walter, L, Gauchenova, K, Kolter, G, Knoll, M, Berthold, C.L, Schneider, G, Pleiss, J, Mueller, M, Pohl, M. | | Deposit date: | 2007-06-25 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Protein Design of Thdp-Dependent Enzymes-Engineering Stereoselectivity.
Chembiochem, 9, 2008
|
|
2VBI
 
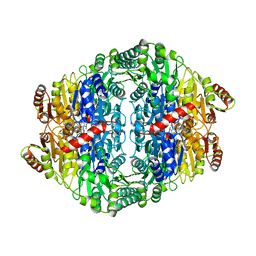 | |
2V9X
 
 | | E138D variant of Escherichia coli dCTP deaminase in complex with dUTP | | Descriptor: | DEOXYCYTIDINE TRIPHOSPHATE DEAMINASE, DEOXYURIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thymark, M, Johansson, E, Larsen, S, Willemoes, M. | | Deposit date: | 2007-08-28 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutational Analysis of the Nucleotide Binding Site of Escherichia Coli Dctp Deaminase.
Arch.Biochem.Biophys., 470, 2008
|
|
2V02
 
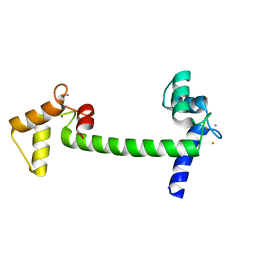 | | Recombinant vertebrate calmodulin complexed with Ba | | Descriptor: | BARIUM ION, CALCIUM ION, CALMODULIN | | Authors: | Kursula, P, Majava, V. | | Deposit date: | 2007-05-04 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Structural Insight Into Lead Neurotoxicity and Calmodulin Activation by Heavy Metals.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2V15
 
 | | Terbium binding in Streptococcus suis Dpr protein | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Havukainen, H, Papageorgiou, A.C, Kauko, A. | | Deposit date: | 2007-05-22 | | Release date: | 2008-06-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the zinc- and terbium-mediated inhibition of ferroxidase activity in Dps ferritin-like proteins.
Protein Sci., 17, 2008
|
|
2UWF
 
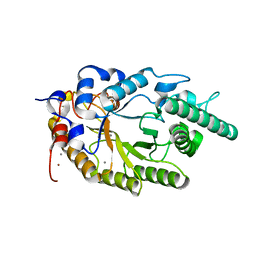 | | Crystal structure of family 10 xylanase from Bacillus halodurans | | Descriptor: | ALKALINE ACTIVE ENDOXYLANASE, CALCIUM ION, COPPER (II) ION | | Authors: | Mamo, G, Thunnissen, M, Hatti-Kaul, R, Mattiasson, B. | | Deposit date: | 2007-03-21 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Alkaline Active Xylanase: Insights Into Mechanisms of High Ph Catalytic Adaptation
Biochimie, 91, 2009
|
|
2V01
 
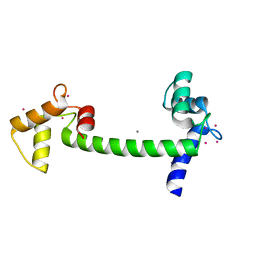 | | Recombinant vertebrate calmodulin complexed with Pb | | Descriptor: | CALCIUM ION, CALMODULIN, LEAD (II) ION | | Authors: | Kursula, P, Majava, V. | | Deposit date: | 2007-05-04 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Structural Insight Into Lead Neurotoxicity and Calmodulin Activation by Heavy Metals.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2V4M
 
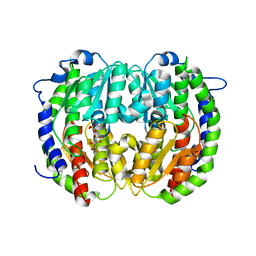 | | The isomerase domain of human glutamine-fructose-6-phosphate transaminase 1 (GFPT1) in complex with fructose 6-phosphate | | Descriptor: | CHLORIDE ION, FRUCTOSE -6-PHOSPHATE, GLUCOSAMINE--FRUCTOSE-6-PHOSPHATE AMINOTRANSFERASE [ISOMERIZING] 1 | | Authors: | Moche, M, Lehtio, L, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Svensson, S, Schueler, H, Thorsell, A.G, Tresaugues, L, Uppenberg, J, Van Den Berg, S, Welin, M, Wisniewska, M, Weigelt, J, Nordlund, P, Wikstrom, M. | | Deposit date: | 2008-09-26 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The Isomerase Domain of Human Gfpt1 in Complex with Fructose 6-Phosphate
To be Published
|
|
2UX1
 
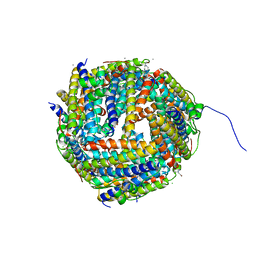 | | Identification of two zinc-binding sites in the Streptococcus suis Dpr protein | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Havukainen, H, Kauko, A, Pulliainen, A.T, Haataja, S, Meyer-Klaucke, W, Finne, J, Papageorgiou, A.C. | | Deposit date: | 2007-03-26 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Zinc- and Terbium-Mediated Inhibition of Ferroxidase Activity in Dps Ferritin- Like Proteins.
Protein Sci., 17, 2008
|
|
2VM8
 
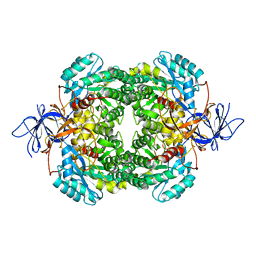 | | Human CRMP-2 crystallised in the presence of Mg | | Descriptor: | DIHYDROPYRIMIDINASE-RELATED PROTEIN 2, MAGNESIUM ION | | Authors: | Kursula, P. | | Deposit date: | 2008-01-24 | | Release date: | 2008-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal and Solution Structure, Stability and Post- Translational Modifications of Collapsin Response Mediator Protein 2.
FEBS J., 275, 2008
|
|
7O46
 
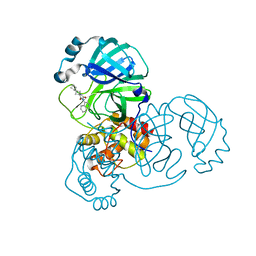 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 17 | | Descriptor: | 2-cyclobutyl-7-isoquinolin-4-yl-5,7-diazaspiro[3.4]octane-6,8-dione, 3C-like proteinase nsp5 | | Authors: | Talibov, V.O. | | Deposit date: | 2021-04-05 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7NYT
 
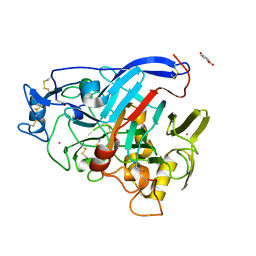 | | Trichoderma reesei Cel7A E212Q mutant in complex with lactose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, Exoglucanase 1, ... | | Authors: | Haataja, T, Sandgren, M, Stahlberg, J. | | Deposit date: | 2021-03-23 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: insights from crystal structures and MD simulation.
Febs J., 290, 2023
|
|
7OC1
 
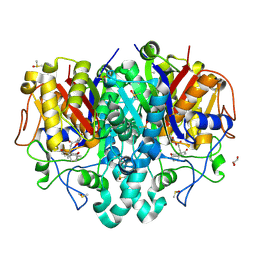 | | Structure of Pseudomonas aeruginosa FabF mutant C164Q in complex with Platensimycin | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Georgiou, C, Brenk, R, Espeland, L.O, Klein, R. | | Deposit date: | 2021-04-25 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Experimental Toolbox for Structure-Based Hit Discovery for P. aeruginosa FabF, a Promising Target for Antibiotics.
Chemmedchem, 16, 2021
|
|
7OC0
 
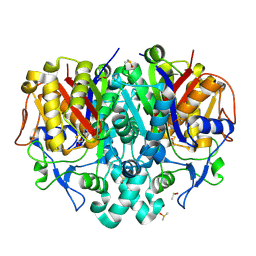 | | Structure of Pseudomonas aeruginosa FabF mutant C164Q in complex with a ligand (2S,4R)-2-(thiophen-2-yl)thiazolidine-4-carboxylic acid | | Descriptor: | (2S,4R)-2-(thiophen-2-yl)thiazolidine-4-carboxylic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Georgiou, C, Brenk, R, Espeland, L.O, Klein, R. | | Deposit date: | 2021-04-25 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | An Experimental Toolbox for Structure-Based Hit Discovery for P. aeruginosa FabF, a Promising Target for Antibiotics.
Chemmedchem, 16, 2021
|
|
