1JXZ
 
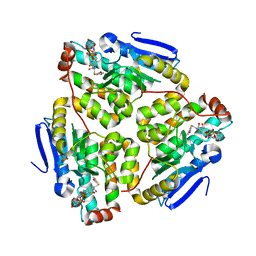 | | Structure of the H90Q mutant of 4-Chlorobenzoyl-Coenzyme A Dehalogenase complexed with 4-hydroxybenzoyl-Coenzyme A (product) | | Descriptor: | 4-HYDROXYBENZOYL COENZYME A, 4-chlorobenzoyl Coenzyme A dehalogenase, CALCIUM ION, ... | | Authors: | Thoden, J.B, Zhang, W, Wei, Y, Luo, L, Taylor, K.L, Yang, G, Dunaway-Mariano, D, Benning, M.M, Holden, H.M. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Histidine 90 Function in 4-chlorobenzoyl-coenzyme A Dehalogenase Catalysis
Biochemistry, 40, 2001
|
|
1JZ8
 
 | |
1JU3
 
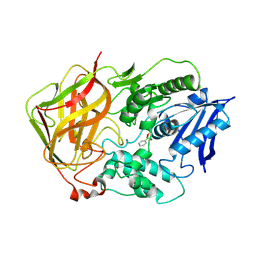 | | BACTERIAL COCAINE ESTERASE COMPLEX WITH TRANSITION STATE ANALOG | | Descriptor: | PHENYL BORONIC ACID, cocaine esterase | | Authors: | Larsen, N.A, Turner, J.M, Stevens, J, Rosser, S.J, Basran, A, Lerner, R.A, Bruce, N.C, Wilson, I.A. | | Deposit date: | 2001-08-23 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of a bacterial cocaine esterase.
Nat.Struct.Biol., 9, 2002
|
|
1K0Y
 
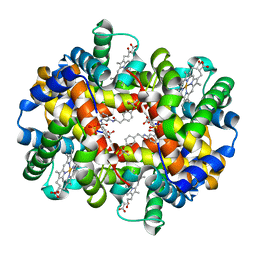 | | X-ray Crystallographic Analyses of Symmetrical Allosteric Effectors of Hemoglobin. Compounds Designed to Link Primary and Secondary Binding Sites | | Descriptor: | 2-{4-[(3{2-[4-(1-CARBOXY-1-METHYL-ETHOXY)-PHENYL]-ACETYLAMINO}-PHENYLCARBAMOYL)-METHYL]-PHENOXY}-2-METHYL-PROPIONIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Safo, M.K, Boyiri, T, Burnett, J.C, Danso-Danquah, R, Moure, C.M, Joshi, G.S, Abraham, D.J. | | Deposit date: | 2001-09-21 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | X-ray crystallographic analyses of symmetrical allosteric effectors of hemoglobin: compounds designed to link primary and secondary binding sites.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1B9J
 
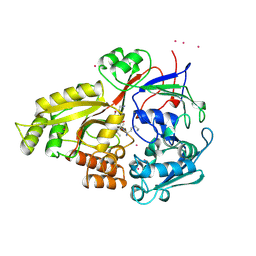 | |
7VVR
 
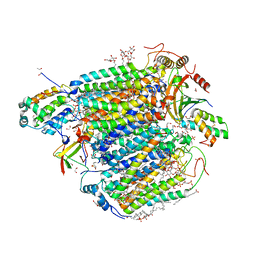 | | Bovine cytochrome c oxidese in CN-bound mixed valence state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Tsukihara, T. | | Deposit date: | 2021-11-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
7VR8
 
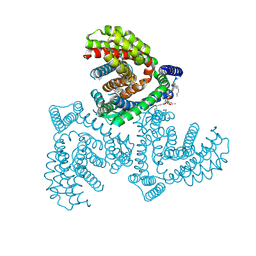 | | Inward-facing structure of human EAAT2 in the substrate-free state | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | Authors: | Kato, T, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2.
Nat Commun, 13, 2022
|
|
7VUW
 
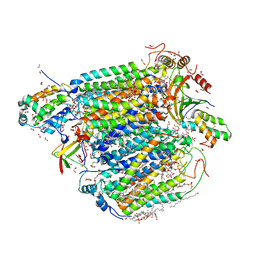 | | Bovine heart cytochrome c oxidase in the cyanide-bound fully oxidized state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Tsukihara, T. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
7VVY
 
 | | TRA module of NuA4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Chen, Z, Qu, K. | | Deposit date: | 2021-11-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the NuA4 acetyltransferase complex bound to the nucleosome.
Nature, 610, 2022
|
|
1BD9
 
 | |
7VVZ
 
 | | NuA4 bound to the nucleosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Qu, K, Chen, Z. | | Deposit date: | 2021-11-09 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structure of the NuA4 acetyltransferase complex bound to the nucleosome.
Nature, 610, 2022
|
|
7VWN
 
 | | The structure of an engineered PET hydrolase | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase | | Authors: | Xie, W, Jia, Q. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An engineered PET hydrolase for biodegradation of microplastics in ocean water
To Be Published
|
|
1BG5
 
 | | CRYSTAL STRUCTURE OF THE ANKYRIN BINDING DOMAIN OF ALPHA-NA,K-ATPASE AS A FUSION PROTEIN WITH GLUTATHIONE S-TRANSFERASE | | Descriptor: | FUSION PROTEIN OF ALPHA-NA,K-ATPASE WITH GLUTATHIONE S-TRANSFERASE | | Authors: | Zhang, Z, Devarajan, P, Morrow, J.S. | | Deposit date: | 1998-06-05 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the ankyrin-binding domain of alpha-Na,K-ATPase.
J.Biol.Chem., 273, 1998
|
|
7VR7
 
 | | Inward-facing structure of human EAAT2 in the WAY213613-bound state | | Descriptor: | (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]amino]-4-oxidanylidene-butanoic acid, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Kato, T, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2.
Nat Commun, 13, 2022
|
|
7VFJ
 
 | | Cytochrome c-type biogenesis protein CcmABCD | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Zhu, J.P, Zhang, K, Li, J, Zheng, W, Gu, M. | | Deposit date: | 2021-09-13 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7VEW
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with unsaturated trigalacturonic acid | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEQ
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in an open conformation | | Descriptor: | GLYCEROL, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VFP
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with heme and single ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-09-13 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7VET
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a closed conformation | | Descriptor: | SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEV
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
1BDJ
 
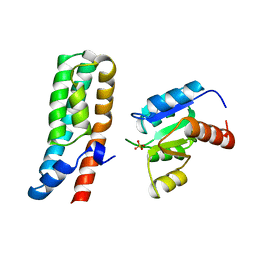 | | COMPLEX STRUCTURE OF HPT DOMAIN AND CHEY | | Descriptor: | AEROBIC RESPIRATION CONTROL SENSOR PROTEIN ARCB, CHEY, SULFATE ION | | Authors: | Kato, M, Mizuno, T, Shimizu, T, Hakoshima, T. | | Deposit date: | 1998-05-10 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure of the histidine-containing phosphotransfer (HPt) domain of the anaerobic sensor protein ArcB complexed with the chemotaxis response regulator CheY.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7VOV
 
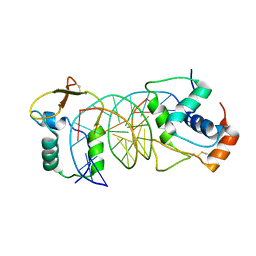 | | The crystal structure of human forkhead box protein in complex with DNA 2 | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*AP*TP*TP*TP*AP*TP*TP*AP*TP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*AP*TP*AP*AP*TP*AP*AP*AP*TP*AP*TP*T)-3'), Forkhead box protein L2 | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
7VXS
 
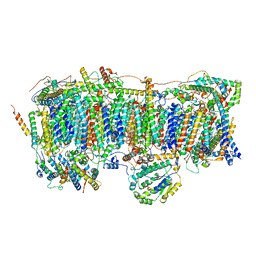 | | Membrane arm of active state CI from Q10 dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-13 | | Release date: | 2022-11-23 | | Last modified: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VER
 
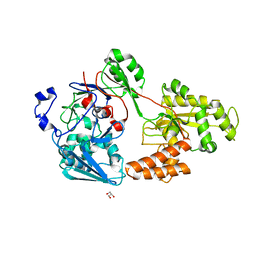 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a full open conformation | | Descriptor: | GLYCEROL, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
1BA5
 
 | | DNA-BINDING DOMAIN OF HUMAN TELOMERIC PROTEIN, HTRF1, NMR, 18 STRUCTURES | | Descriptor: | HTRF1 | | Authors: | Nishikawa, T, Nagadoi, A, Yoshimura, S, Aimoto, S, Nishimura, Y. | | Deposit date: | 1998-04-22 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of human telomeric protein, hTRF1.
Structure, 6, 1998
|
|
