8QA1
 
 | |
8Q6X
 
 | | Crystal structure of Cytochrome P450 GymB5 from Streptomyces katrae | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Freytag, J, Mueller, J.M, Stehle, T, Zocher, G. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Intramolecular Coupling and Nucleobase Transfer - How Cytochrome P450 Enzymes GymBx Establish Their Chemoselectivity
Chemcatchem, 16, 2024
|
|
5KIC
 
 | | Long-sought stabilization of berkelium(IV) in solution: An anomaly within the heavy actinide series | | Descriptor: | CALIFORNIUM ION, CHLORIDE ION, N,N'-butane-1,4-diylbis[1-hydroxy-N-(3-{[(1-hydroxy-6-oxo-1,6-dihydropyridin-2-yl)carbonyl]amino}propyl)-6-oxo-1,6-dihydropyridine-2-carboxamide], ... | | Authors: | Rupert, P.B, Strong, R.K. | | Deposit date: | 2016-06-16 | | Release date: | 2017-04-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chelation and stabilization of berkelium in oxidation state +IV.
Nat Chem, 9, 2017
|
|
8Q6Y
 
 | | Crystal structure of Cytochrome P450 GymB5 from Streptomyces katrae in complex with cYY and Hypoxanthine | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, 1,2-ETHANEDIOL, Cytochrome P450, ... | | Authors: | Freytag, J, Mueller, J.M, Stehle, T, Zocher, G. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Intramolecular Coupling and Nucleobase Transfer - How Cytochrome P450 Enzymes GymBx Establish Their Chemoselectivity
Chemcatchem, 16, 2024
|
|
8QA3
 
 | |
8QA0
 
 | |
5K2N
 
 | |
5GVT
 
 | |
8PT5
 
 | | ERK2 covelently bound to RU187 cyclohexenone based inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sok, P, Poti, A, Remenyi, A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
8PMF
 
 | |
7QQ6
 
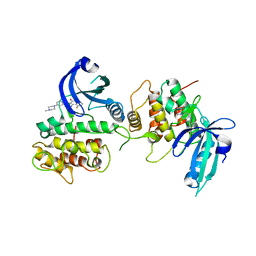 | | GCN2 (EIF2ALPHA KINASE 4, E2AK4) IN COMPLEX WITH COMPOUND 1 (dovitinib) | | Descriptor: | 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, eIF-2-alpha kinase GCN2 | | Authors: | Maia de Oliveira, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of human GCN2 reveals a parallel, back-to-back kinase dimer with a plastic DFG activation loop motif.
Biochem.J., 477, 2020
|
|
5K5J
 
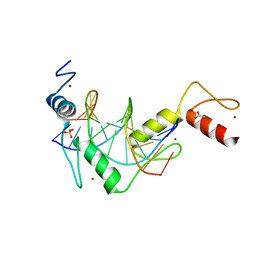 | |
7Q8V
 
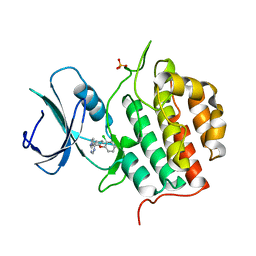 | | Crystal structure of TTBK1 in complex with VNG2.73 (compound 42) | | Descriptor: | PHOSPHATE ION, Tau-tubulin kinase 1, ~{N}-[4-(2-chloranylphenoxy)phenyl]-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
8PR7
 
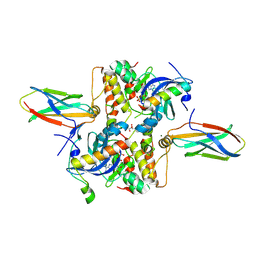 | |
5K7O
 
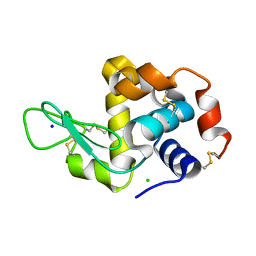 | | MicroED structure of lysozyme at 1.8 A resolution | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.8 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
8PSY
 
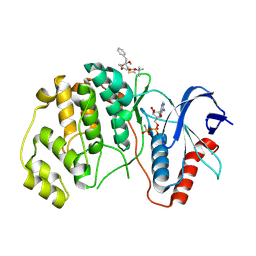 | | ERK2 covelently bound to RU68 cyclohexenone based inhibitor | | Descriptor: | AMP PHOSPHORAMIDATE, Mitogen-activated protein kinase 1, ~{O}3-~{tert}-butyl ~{O}1-methyl (1~{S},3~{R})-4-oxidanylidene-1-(phenylmethyl)cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
8PSW
 
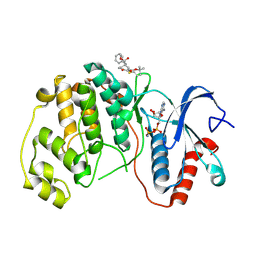 | | ERK2 covalently bound to RU67 cyclohexenone based inhibitor | | Descriptor: | AMP PHOSPHORAMIDATE, Mitogen-activated protein kinase 1, ~{O}3-~{tert}-butyl ~{O}1-methyl (1~{R},3~{R})-4-oxidanylidene-1-(phenylmethyl)cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
7QPZ
 
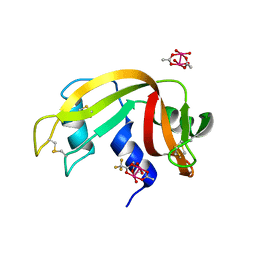 | |
8PT1
 
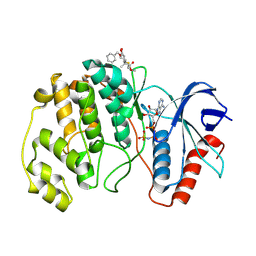 | | ERK2 covelently bound to RU76 cyclohexenone based inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ~{O}1-methyl ~{O}3-(phenylmethyl) (1~{S},3~{R})-1-methyl-4-oxidanylidene-cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
7QQ1
 
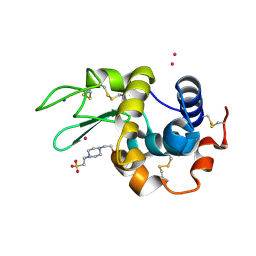 | |
7Q8Y
 
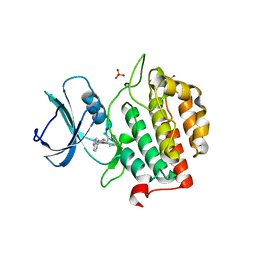 | | Crystal structure of TTBK2 in complex with VNG2.73 (compound 42) | | Descriptor: | PHOSPHATE ION, Tau-tubulin kinase 2, ~{N}-[4-(2-chloranylphenoxy)phenyl]-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
7Q8Z
 
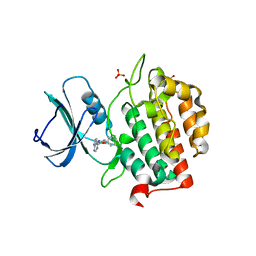 | | Crystal structure of TTBK2 in complex with VNG1.33 (compound 27) | | Descriptor: | PHOSPHATE ION, Tau-tubulin kinase 2, ~{N}-(4-phenoxyphenyl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
5K2P
 
 | | Crystal structure of lysozyme | | Descriptor: | Lysozyme C | | Authors: | Ko, S, Choe, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of lysozyme
To Be Published
|
|
8PVX
 
 | |
8PTA
 
 | | JNK1 covalently bound to BD837 cyclohexenone based inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 8, methyl (1R,3S)-1-methyl-3-[[3-[[3-methyl-4-[(4-pyridin-3-ylpyrimidin-2-yl)amino]phenyl]carbamoyl]phenyl]carbamoyl]-4-oxidanylidene-cyclohexane-1-carboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Tunable c-Jun N-terminal kinase (JNK) inhibitors that target a specific cysteine by a reversible covalent bond
To Be Published
|
|
